| Entry | Database: PDB / ID: 5t03
|
|---|
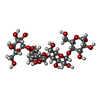
| Title | Crystal structure of heparan sulfate 6-O-sulfotransferase with bound PAP and glucuronic acid containing hexasaccharide substrate |
|---|
 Components Components | maltose binding protein - heparan sulfate 6-O-sulfotransferase isoform 3 fusion protein |
|---|
 Keywords Keywords | TRANSFERASE / heparan sulfate / sulfotransferase / complex |
|---|
| Function / homology |  Function and homology information Function and homology information
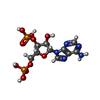
heparan sulfate 6-sulfotransferase activity / HS-GAG biosynthesis / Transferases; Transferring sulfur-containing groups; Sulfotransferases / 3'-phosphoadenosine 5'-phosphosulfate binding / sulfotransferase activity / heparan sulfate proteoglycan biosynthetic process / oligosaccharide binding / detection of maltose stimulus / maltose transport complex / carbohydrate transport ...heparan sulfate 6-sulfotransferase activity / HS-GAG biosynthesis / Transferases; Transferring sulfur-containing groups; Sulfotransferases / 3'-phosphoadenosine 5'-phosphosulfate binding / sulfotransferase activity / heparan sulfate proteoglycan biosynthetic process / oligosaccharide binding / detection of maltose stimulus / maltose transport complex / carbohydrate transport / carbohydrate transmembrane transporter activity / maltose binding / maltose transport / maltodextrin transmembrane transport / ATP-binding cassette (ABC) transporter complex, substrate-binding subunit-containing / ATP-binding cassette (ABC) transporter complex / cell chemotaxis / outer membrane-bounded periplasmic space / periplasmic space / DNA damage response / membraneSimilarity search - Function Heparan sulphate 6-sulfotransferase/Protein-tyrosine sulfotransferase / Sulfotransferase / Sulfotransferase family / Maltose/Cyclodextrin ABC transporter, substrate-binding protein / Solute-binding family 1, conserved site / Bacterial extracellular solute-binding proteins, family 1 signature. / Bacterial extracellular solute-binding protein / Bacterial extracellular solute-binding protein / Periplasmic binding protein-like II / D-Maltodextrin-Binding Protein; domain 2 ...Heparan sulphate 6-sulfotransferase/Protein-tyrosine sulfotransferase / Sulfotransferase / Sulfotransferase family / Maltose/Cyclodextrin ABC transporter, substrate-binding protein / Solute-binding family 1, conserved site / Bacterial extracellular solute-binding proteins, family 1 signature. / Bacterial extracellular solute-binding protein / Bacterial extracellular solute-binding protein / Periplasmic binding protein-like II / D-Maltodextrin-Binding Protein; domain 2 / P-loop containing nucleotide triphosphate hydrolases / Rossmann fold / P-loop containing nucleoside triphosphate hydrolase / 3-Layer(aba) Sandwich / Alpha BetaSimilarity search - Domain/homology alpha-maltotetraose / ADENOSINE-3'-5'-DIPHOSPHATE / P-NITROPHENOL / L(+)-TARTARIC ACID / Heparan-sulfate 6-O-sulfotransferase 3-B / Maltose/maltodextrin-binding periplasmic protein / Maltose/maltodextrin-binding periplasmic proteinSimilarity search - Component |
|---|
| Biological species |   Escherichia coli O157:H7 (bacteria) Escherichia coli O157:H7 (bacteria)
  Danio rerio (zebrafish) Danio rerio (zebrafish) |
|---|
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.1 Å MOLECULAR REPLACEMENT / Resolution: 2.1 Å |
|---|
 Authors Authors | Pedersen, L.C. / Moon, A.F. / Krahn, J.M. / Liu, J. |
|---|
| Funding support |  United States, 2items United States, 2items | Organization | Grant number | Country |
|---|
| National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS) | GM102137 |  United States United States | | National Institutes of Health/National Heart, Lung, and Blood Institute (NIH/NHLBI) | HL094463 |  United States United States |
|
|---|
 Citation Citation |  Journal: ACS Chem. Biol. / Year: 2017 Journal: ACS Chem. Biol. / Year: 2017
Title: Structure Based Substrate Specificity Analysis of Heparan Sulfate 6-O-Sulfotransferases.
Authors: Xu, Y. / Moon, A.F. / Xu, S. / Krahn, J.M. / Liu, J. / Pedersen, L.C. |
|---|
| History | | Deposition | Aug 15, 2016 | Deposition site: RCSB / Processing site: RCSB |
|---|
| Revision 1.0 | Feb 1, 2017 | Provider: repository / Type: Initial release |
|---|
| Revision 1.1 | Sep 20, 2017 | Group: Author supporting evidence / Category: pdbx_audit_support / Item: _pdbx_audit_support.funding_organization |
|---|
| Revision 1.2 | Dec 4, 2019 | Group: Author supporting evidence / Data collection / Category: chem_comp / pdbx_audit_support
Item: _chem_comp.type / _pdbx_audit_support.funding_organization |
|---|
| Revision 2.0 | Jul 29, 2020 | Group: Atomic model / Data collection ...Atomic model / Data collection / Derived calculations / Structure summary
Category: atom_site / chem_comp ...atom_site / chem_comp / entity / entity_name_com / pdbx_branch_scheme / pdbx_chem_comp_identifier / pdbx_entity_branch / pdbx_entity_branch_descriptor / pdbx_entity_branch_link / pdbx_entity_branch_list / pdbx_entity_nonpoly / pdbx_molecule_features / pdbx_nonpoly_scheme / pdbx_struct_assembly_gen / pdbx_struct_conn_angle / struct_asym / struct_conn / struct_conn_type / struct_site / struct_site_gen
Item: _atom_site.B_iso_or_equiv / _atom_site.Cartn_x ..._atom_site.B_iso_or_equiv / _atom_site.Cartn_x / _atom_site.Cartn_y / _atom_site.Cartn_z / _atom_site.auth_asym_id / _atom_site.auth_atom_id / _atom_site.auth_comp_id / _atom_site.auth_seq_id / _atom_site.label_alt_id / _atom_site.label_asym_id / _atom_site.label_atom_id / _atom_site.label_comp_id / _atom_site.label_entity_id / _atom_site.occupancy / _atom_site.type_symbol / _chem_comp.mon_nstd_flag / _chem_comp.name / _chem_comp.type / _pdbx_struct_assembly_gen.asym_id_list / _pdbx_struct_conn_angle.ptnr1_auth_asym_id / _pdbx_struct_conn_angle.ptnr1_auth_comp_id / _pdbx_struct_conn_angle.ptnr1_auth_seq_id / _pdbx_struct_conn_angle.ptnr1_label_asym_id / _pdbx_struct_conn_angle.ptnr1_label_atom_id / _pdbx_struct_conn_angle.ptnr1_label_comp_id / _pdbx_struct_conn_angle.ptnr1_label_seq_id / _pdbx_struct_conn_angle.ptnr2_auth_asym_id / _pdbx_struct_conn_angle.ptnr2_auth_seq_id / _pdbx_struct_conn_angle.ptnr2_label_asym_id / _pdbx_struct_conn_angle.ptnr3_auth_asym_id / _pdbx_struct_conn_angle.ptnr3_auth_comp_id / _pdbx_struct_conn_angle.ptnr3_auth_seq_id / _pdbx_struct_conn_angle.ptnr3_label_asym_id / _pdbx_struct_conn_angle.ptnr3_label_atom_id / _pdbx_struct_conn_angle.ptnr3_label_comp_id / _pdbx_struct_conn_angle.ptnr3_label_seq_id / _pdbx_struct_conn_angle.value / _struct_conn.conn_type_id / _struct_conn.id / _struct_conn.pdbx_dist_value / _struct_conn.pdbx_leaving_atom_flag / _struct_conn.pdbx_ptnr1_label_alt_id / _struct_conn.pdbx_ptnr2_label_alt_id / _struct_conn.ptnr1_auth_asym_id / _struct_conn.ptnr1_auth_comp_id / _struct_conn.ptnr1_auth_seq_id / _struct_conn.ptnr1_label_asym_id / _struct_conn.ptnr1_label_atom_id / _struct_conn.ptnr1_label_comp_id / _struct_conn.ptnr1_label_seq_id / _struct_conn.ptnr2_auth_asym_id / _struct_conn.ptnr2_auth_comp_id / _struct_conn.ptnr2_auth_seq_id / _struct_conn.ptnr2_label_asym_id / _struct_conn.ptnr2_label_atom_id / _struct_conn.ptnr2_label_comp_id / _struct_conn_type.id
Description: Carbohydrate remediation / Provider: repository / Type: Remediation |
|---|
| Revision 2.1 | Oct 23, 2024 | Group: Data collection / Database references ...Data collection / Database references / Derived calculations / Structure summary
Category: chem_comp / chem_comp_atom ...chem_comp / chem_comp_atom / chem_comp_bond / database_2 / pdbx_entry_details / pdbx_modification_feature / struct_conn
Item: _chem_comp.pdbx_synonyms / _database_2.pdbx_DOI ..._chem_comp.pdbx_synonyms / _database_2.pdbx_DOI / _database_2.pdbx_database_accession / _struct_conn.pdbx_leaving_atom_flag |
|---|
|
|---|
 Yorodumi
Yorodumi Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information

 X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON /
SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.1 Å
MOLECULAR REPLACEMENT / Resolution: 2.1 Å  Authors
Authors United States, 2items
United States, 2items  Citation
Citation Journal: ACS Chem. Biol. / Year: 2017
Journal: ACS Chem. Biol. / Year: 2017 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 5t03.cif.gz
5t03.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb5t03.ent.gz
pdb5t03.ent.gz PDB format
PDB format 5t03.json.gz
5t03.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/t0/5t03
https://data.pdbj.org/pub/pdb/validation_reports/t0/5t03 ftp://data.pdbj.org/pub/pdb/validation_reports/t0/5t03
ftp://data.pdbj.org/pub/pdb/validation_reports/t0/5t03 Links
Links Assembly
Assembly


 Components
Components














 X-RAY DIFFRACTION / Number of used crystals: 1
X-RAY DIFFRACTION / Number of used crystals: 1  Sample preparation
Sample preparation SYNCHROTRON / Site:
SYNCHROTRON / Site:  APS
APS  / Beamline: 22-ID / Wavelength: 1 Å
/ Beamline: 22-ID / Wavelength: 1 Å Processing
Processing MOLECULAR REPLACEMENT / Resolution: 2.1→38.648 Å / SU ML: 0.19 / Cross valid method: THROUGHOUT / σ(F): 1.36 / Phase error: 20.88 / Stereochemistry target values: ML
MOLECULAR REPLACEMENT / Resolution: 2.1→38.648 Å / SU ML: 0.19 / Cross valid method: THROUGHOUT / σ(F): 1.36 / Phase error: 20.88 / Stereochemistry target values: ML Movie
Movie Controller
Controller














 PDBj
PDBj