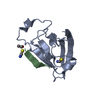
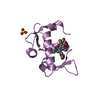
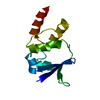
Entry Database : PDB / ID : 5ovpTitle PDZ domain from rat Shank3 bound to the C terminus of CIRL Adhesion G protein-coupled receptor L1 SH3 and multiple ankyrin repeat domains protein 3 Keywords / / / / Function / homology Function Domain/homology Component
/ / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / Biological species Rattus norvegicus (Norway rat)Method / / Resolution : 1.5 Å Authors Ponna, S.K. / Myllykoski, M. / Boeckers, T.M. / Kursula, P. Journal : J. Neurochem. / Year : 2018Title : Structural basis for PDZ domain interactions in the post-synaptic density scaffolding protein Shank3.Authors : Ponna, S.K. / Ruskamo, S. / Myllykoski, M. / Keller, C. / Boeckers, T.M. / Kursula, P. History Deposition Aug 29, 2017 Deposition site / Processing site Revision 1.0 Mar 7, 2018 Provider / Type Revision 1.1 Jul 11, 2018 Group / Database references / Category Item / _citation.page_first / _citation.page_lastRevision 1.2 Oct 23, 2024 Group / Database references / Structure summaryCategory chem_comp_atom / chem_comp_bond ... chem_comp_atom / chem_comp_bond / database_2 / pdbx_entry_details / pdbx_modification_feature Item / _database_2.pdbx_database_accession
Show all Show less
 Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information
 X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON / Resolution: 1.5 Å
SYNCHROTRON / Resolution: 1.5 Å  Authors
Authors Citation
Citation Journal: J. Neurochem. / Year: 2018
Journal: J. Neurochem. / Year: 2018 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 5ovp.cif.gz
5ovp.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb5ovp.ent.gz
pdb5ovp.ent.gz PDB format
PDB format 5ovp.json.gz
5ovp.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/ov/5ovp
https://data.pdbj.org/pub/pdb/validation_reports/ov/5ovp ftp://data.pdbj.org/pub/pdb/validation_reports/ov/5ovp
ftp://data.pdbj.org/pub/pdb/validation_reports/ov/5ovp Links
Links Assembly
Assembly
 Components
Components


 X-RAY DIFFRACTION / Number of used crystals: 1
X-RAY DIFFRACTION / Number of used crystals: 1  Sample preparation
Sample preparation SYNCHROTRON / Site:
SYNCHROTRON / Site:  PETRA III, EMBL c/o DESY
PETRA III, EMBL c/o DESY  / Beamline: P13 (MX1) / Wavelength: 1 Å
/ Beamline: P13 (MX1) / Wavelength: 1 Å Processing
Processing Movie
Movie Controller
Controller














 PDBj
PDBj


