+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 | データベース: PDB / ID: 5b1b | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| タイトル | Bovine heart cytochrome c oxidase in the fully reduced state at 1.6 angstrom resolution | ||||||||||||
 要素 要素 | (Cytochrome c oxidase subunit ...) x 13 | ||||||||||||
 キーワード キーワード | OXIDOREDUCTASE / respiratory chain / Proton pump / Heme | ||||||||||||
| 機能・相同性 |  機能・相同性情報 機能・相同性情報Complex IV assembly / TP53 Regulates Metabolic Genes / respiratory chain complex IV assembly / Cytoprotection by HMOX1 / mitochondrial respirasome assembly / Respiratory electron transport / respiratory chain complex IV / respiratory chain complex / cytochrome-c oxidase / oxidative phosphorylation ...Complex IV assembly / TP53 Regulates Metabolic Genes / respiratory chain complex IV assembly / Cytoprotection by HMOX1 / mitochondrial respirasome assembly / Respiratory electron transport / respiratory chain complex IV / respiratory chain complex / cytochrome-c oxidase / oxidative phosphorylation / mitochondrial electron transport, cytochrome c to oxygen / cytochrome-c oxidase activity / Mitochondrial protein degradation / ATP synthesis coupled electron transport / enzyme regulator activity / aerobic respiration / central nervous system development / respiratory electron transport chain / oxidoreductase activity / mitochondrial inner membrane / copper ion binding / heme binding / mitochondrion / metal ion binding 類似検索 - 分子機能 | ||||||||||||
| 生物種 |  | ||||||||||||
| 手法 |  X線回折 / X線回折 /  シンクロトロン / シンクロトロン /  分子置換 / 解像度: 1.6 Å 分子置換 / 解像度: 1.6 Å | ||||||||||||
 データ登録者 データ登録者 | Yano, N. / Muramoto, K. / Shimada, A. / Takemura, S. / Baba, J. / Fujisawa, H. / Mochizuki, M. / Shinzawa-Itoh, K. / Yamashita, E. / Tsukihara, T. / Yoshikawa, S. | ||||||||||||
| 資金援助 |  日本, 3件 日本, 3件
| ||||||||||||
 引用 引用 |  ジャーナル: J.Biol.Chem. / 年: 2016 ジャーナル: J.Biol.Chem. / 年: 2016タイトル: The Mg2+-containing Water Cluster of Mammalian Cytochrome c Oxidase Collects Four Pumping Proton Equivalents in Each Catalytic Cycle. 著者: Yano, N. / Muramoto, K. / Shimada, A. / Takemura, S. / Baba, J. / Fujisawa, H. / Mochizuki, M. / Shinzawa-Itoh, K. / Yamashita, E. / Tsukihara, T. / Yoshikawa, S. | ||||||||||||
| 履歴 |
|
- 構造の表示
構造の表示
| 構造ビューア | 分子:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- ダウンロードとリンク
ダウンロードとリンク
- ダウンロード
ダウンロード
| PDBx/mmCIF形式 |  5b1b.cif.gz 5b1b.cif.gz | 1.5 MB | 表示 |  PDBx/mmCIF形式 PDBx/mmCIF形式 |
|---|---|---|---|---|
| PDB形式 |  pdb5b1b.ent.gz pdb5b1b.ent.gz | 1.3 MB | 表示 |  PDB形式 PDB形式 |
| PDBx/mmJSON形式 |  5b1b.json.gz 5b1b.json.gz | ツリー表示 |  PDBx/mmJSON形式 PDBx/mmJSON形式 | |
| その他 |  その他のダウンロード その他のダウンロード |
-検証レポート
| 文書・要旨 |  5b1b_validation.pdf.gz 5b1b_validation.pdf.gz | 13 MB | 表示 |  wwPDB検証レポート wwPDB検証レポート |
|---|---|---|---|---|
| 文書・詳細版 |  5b1b_full_validation.pdf.gz 5b1b_full_validation.pdf.gz | 13.1 MB | 表示 | |
| XML形式データ |  5b1b_validation.xml.gz 5b1b_validation.xml.gz | 188.1 KB | 表示 | |
| CIF形式データ |  5b1b_validation.cif.gz 5b1b_validation.cif.gz | 258.8 KB | 表示 | |
| アーカイブディレクトリ |  https://data.pdbj.org/pub/pdb/validation_reports/b1/5b1b https://data.pdbj.org/pub/pdb/validation_reports/b1/5b1b ftp://data.pdbj.org/pub/pdb/validation_reports/b1/5b1b ftp://data.pdbj.org/pub/pdb/validation_reports/b1/5b1b | HTTPS FTP |
-関連構造データ
- リンク
リンク
- 集合体
集合体
| 登録構造単位 | 
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | 
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 単位格子 |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 非結晶学的対称性 (NCS) | NCS oper:
|
- 要素
要素
-Cytochrome c oxidase subunit ... , 13種, 26分子 ANBOCPDQERFSGTHUIVJWKXLYMZ
| #1: タンパク質 | 分子量: 57093.852 Da / 分子数: 2 / 由来タイプ: 天然 / 由来: (天然)  #2: タンパク質 | 分子量: 26068.404 Da / 分子数: 2 / 由来タイプ: 天然 / 由来: (天然)  #3: タンパク質 | 分子量: 29725.328 Da / 分子数: 2 / 由来タイプ: 天然 / 由来: (天然)  #4: タンパク質 | 分子量: 16913.367 Da / 分子数: 2 / 由来タイプ: 天然 / 由来: (天然)  #5: タンパク質 | 分子量: 12083.727 Da / 分子数: 2 / 由来タイプ: 天然 / 由来: (天然)  #6: タンパク質 | 分子量: 10684.038 Da / 分子数: 2 / 由来タイプ: 天然 / 由来: (天然)  #7: タンパク質 | 分子量: 9452.687 Da / 分子数: 2 / 由来タイプ: 天然 / 由来: (天然)  #8: タンパク質 | 分子量: 9411.600 Da / 分子数: 2 / 由来タイプ: 天然 / 由来: (天然)  #9: タンパク質 | 分子量: 8537.019 Da / 分子数: 2 / 由来タイプ: 天然 / 由来: (天然)  #10: タンパク質 | 分子量: 6553.546 Da / 分子数: 2 / 由来タイプ: 天然 / 由来: (天然)  #11: タンパク質・ペプチド | 分子量: 5442.168 Da / 分子数: 2 / 由来タイプ: 天然 / 由来: (天然)  #12: タンパク質・ペプチド | 分子量: 5362.319 Da / 分子数: 2 / 由来タイプ: 天然 / 由来: (天然)  #13: タンパク質・ペプチド | 分子量: 4738.523 Da / 分子数: 2 / 由来タイプ: 天然 / 由来: (天然)  |
|---|
-糖 , 1種, 23分子 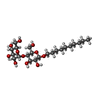
| #24: 糖 | ChemComp-DMU / |
|---|
-非ポリマー , 15種, 3413分子 








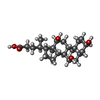



















| #14: 化合物 | ChemComp-HEA / #15: 化合物 | #16: 化合物 | #17: 化合物 | #18: 化合物 | ChemComp-PGV / ( #19: 化合物 | ChemComp-TGL / #20: 化合物 | #21: 化合物 | ChemComp-EDO / #22: 化合物 | #23: 化合物 | ChemComp-CHD / #25: 化合物 | ChemComp-CDL / #26: 化合物 | ChemComp-PEK / ( #27: 化合物 | #28: 化合物 | #29: 水 | ChemComp-HOH / | |
|---|
-詳細
| 研究の焦点であるリガンドがあるか | N |
|---|
-実験情報
-実験
| 実験 | 手法:  X線回折 X線回折 |
|---|
- 試料調製
試料調製
| 結晶 | マシュー密度: 4.01 Å3/Da / 溶媒含有率: 69.36 % |
|---|---|
| 結晶化 | 温度: 277 K / 手法: batch mode / pH: 6.8 / 詳細: PEG 4000, Sodium phosphate, Decylmaltoside |
-データ収集
| 回折 | 平均測定温度: 50 K / Serial crystal experiment: N |
|---|---|
| 放射光源 | 由来:  シンクロトロン / サイト: シンクロトロン / サイト:  SPring-8 SPring-8  / ビームライン: BL44XU / 波長: 0.9 Å / ビームライン: BL44XU / 波長: 0.9 Å |
| 検出器 | タイプ: RAYONIX MX225HE / 検出器: CCD / 日付: 2010年4月23日 |
| 放射 | プロトコル: SINGLE WAVELENGTH / 単色(M)・ラウエ(L): M / 散乱光タイプ: x-ray |
| 放射波長 | 波長: 0.9 Å / 相対比: 1 |
| 反射 | 解像度: 1.6→200 Å / Num. obs: 850991 / % possible obs: 99.6 % / 冗長度: 6.7 % / Rmerge(I) obs: 0.116 / Net I/σ(I): 25.5 |
| 反射 シェル | 解像度: 1.6→1.62 Å / 冗長度: 5.4 % / Rmerge(I) obs: 0.74 / Num. unique obs: 28158 / % possible all: 99.4 |
- 解析
解析
| ソフトウェア |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 精密化 | 構造決定の手法:  分子置換 分子置換開始モデル: 2DYR 解像度: 1.6→39.897 Å / SU ML: 0.13 / 交差検証法: FREE R-VALUE / σ(F): 2 / 位相誤差: 20.35 / 立体化学のターゲット値: ML
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 溶媒の処理 | 減衰半径: 0.9 Å / VDWプローブ半径: 1.11 Å / 溶媒モデル: FLAT BULK SOLVENT MODEL | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 精密化ステップ | サイクル: LAST / 解像度: 1.6→39.897 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 拘束条件 |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS精密化 シェル |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 精密化 TLS | 手法: refined / Refine-ID: X-RAY DIFFRACTION
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 精密化 TLSグループ |
|
 ムービー
ムービー コントローラー
コントローラー


























 PDBj
PDBj


















