+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 4gy2 | ||||||
|---|---|---|---|---|---|---|---|
| Title | Crystal structure of apo-Ia-actin complex | ||||||
 Components Components |
| ||||||
 Keywords Keywords | TOXIN/STRUCTURAL PROTEIN / ADP-ribosyltransferase / TOXIN-STRUCTURAL PROTEIN complex | ||||||
| Function / homology |  Function and homology information Function and homology informationcytoskeletal motor activator activity / myosin heavy chain binding / tropomyosin binding / actin filament bundle / troponin I binding / filamentous actin / mesenchyme migration / skeletal muscle myofibril / actin filament bundle assembly / striated muscle thin filament ...cytoskeletal motor activator activity / myosin heavy chain binding / tropomyosin binding / actin filament bundle / troponin I binding / filamentous actin / mesenchyme migration / skeletal muscle myofibril / actin filament bundle assembly / striated muscle thin filament / skeletal muscle thin filament assembly / actin monomer binding / skeletal muscle fiber development / stress fiber / titin binding / actin filament polymerization / actin filament / filopodium / Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to facilitate cellular and subcellular movement / calcium-dependent protein binding / lamellipodium / cell body / hydrolase activity / protein domain specific binding / nucleotide binding / calcium ion binding / positive regulation of gene expression / magnesium ion binding / extracellular region / ATP binding / identical protein binding / cytoplasm Similarity search - Function | ||||||
| Biological species |   | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.71 Å MOLECULAR REPLACEMENT / Resolution: 2.71 Å | ||||||
 Authors Authors | Tsurumura, T. / Oda, M. / Nagahama, M. / Tsuge, H. | ||||||
 Citation Citation |  Journal: Proc.Natl.Acad.Sci.USA / Year: 2013 Journal: Proc.Natl.Acad.Sci.USA / Year: 2013Title: Arginine ADP-ribosylation mechanism based on structural snapshots of iota-toxin and actin complex Authors: Tsurumura, T. / Tsumori, Y. / Qiu, H. / Oda, M. / Sakurai, J. / Nagahama, M. / Tsuge, H. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  4gy2.cif.gz 4gy2.cif.gz | 173.3 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb4gy2.ent.gz pdb4gy2.ent.gz | 134 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  4gy2.json.gz 4gy2.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/gy/4gy2 https://data.pdbj.org/pub/pdb/validation_reports/gy/4gy2 ftp://data.pdbj.org/pub/pdb/validation_reports/gy/4gy2 ftp://data.pdbj.org/pub/pdb/validation_reports/gy/4gy2 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  4h03C  4h0tC  4h0vC  4h0xC  4h0yC  3buzS S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
-Protein , 2 types, 2 molecules AB
| #1: Protein | Mass: 48219.180 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   References: UniProt: Q46220, NAD+-protein-arginine ADP-ribosyltransferase |
|---|---|
| #2: Protein | Mass: 41862.613 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   |
-Non-polymers , 5 types, 147 molecules 


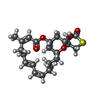





| #3: Chemical | ChemComp-PO4 / |
|---|---|
| #4: Chemical | ChemComp-CA / |
| #5: Chemical | ChemComp-ATP / |
| #6: Chemical | ChemComp-LAR / |
| #7: Water | ChemComp-HOH / |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 3.16 Å3/Da / Density % sol: 61.08 % |
|---|---|
| Crystal grow | Temperature: 277.13 K / Method: vapor diffusion, hanging drop / pH: 6.5 Details: 18% PEG 1500, 0.1M MES, pH 6.5, VAPOR DIFFUSION, HANGING DROP, temperature 277.13K |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  Photon Factory Photon Factory  / Beamline: AR-NW12A / Wavelength: 1 Å / Beamline: AR-NW12A / Wavelength: 1 Å |
| Detector | Type: ADSC QUANTUM 210r / Detector: CCD / Date: Dec 7, 2011 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1 Å / Relative weight: 1 |
| Reflection | Resolution: 2.71→37.19 Å / Num. all: 31974 / Num. obs: 31910 / % possible obs: 99.8 % / Observed criterion σ(F): 0 / Observed criterion σ(I): 0 / Redundancy: 7.1 % / Rmerge(I) obs: 0.098 / Net I/σ(I): 9.9 |
| Reflection shell | Resolution: 2.7→2.75 Å / Redundancy: 6.9 % / Rmerge(I) obs: 0.461 / Mean I/σ(I) obs: 5.1 / Num. unique all: 1539 / % possible all: 99.5 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB ENTRY 3BUZ Resolution: 2.71→37.19 Å / Cor.coef. Fo:Fc: 0.931 / Cor.coef. Fo:Fc free: 0.884 / SU B: 11.672 / SU ML: 0.234 / Cross valid method: THROUGHOUT / ESU R: 0.636 / ESU R Free: 0.326 / Stereochemistry target values: MAXIMUM LIKELIHOOD / Details: HYDROGENS HAVE BEEN USED IF PRESENT IN THE INPUT
| |||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.2 Å / Solvent model: MASK | |||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 33.221 Å2
| |||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.71→37.19 Å
| |||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 2.707→2.777 Å / Total num. of bins used: 20
|
 Movie
Movie Controller
Controller















 PDBj
PDBj






