[English] 日本語
 Yorodumi
Yorodumi- PDB-4bch: Structure of CDK9 in complex with cyclin T and a 2-amino-4-hetero... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 4bch | ||||||
|---|---|---|---|---|---|---|---|
| Title | Structure of CDK9 in complex with cyclin T and a 2-amino-4-heteroaryl- pyrimidine inhibitor | ||||||
 Components Components |
| ||||||
 Keywords Keywords | TRANSFERASE/CELL CYCLE / TRANSFERASE-CELL CYCLE COMPLEX / CDK-CYCLIN COMPLEX / TRANSCRIPTION-PROTEIN BINDING / STRUCTURE-BASED DRUG DESIGN | ||||||
| Function / homology |  Function and homology information Function and homology informationP-TEFb complex / Interactions of Tat with host cellular proteins / nucleus localization / 7SK snRNA binding / cyclin/CDK positive transcription elongation factor complex / regulation of mRNA 3'-end processing / positive regulation of protein localization to chromatin / regulation of muscle cell differentiation / transcription elongation factor activity / regulation of cyclin-dependent protein serine/threonine kinase activity ...P-TEFb complex / Interactions of Tat with host cellular proteins / nucleus localization / 7SK snRNA binding / cyclin/CDK positive transcription elongation factor complex / regulation of mRNA 3'-end processing / positive regulation of protein localization to chromatin / regulation of muscle cell differentiation / transcription elongation factor activity / regulation of cyclin-dependent protein serine/threonine kinase activity / host-mediated activation of viral transcription / cyclin-dependent protein serine/threonine kinase activator activity / RNA polymerase binding / positive regulation of DNA-templated transcription, elongation / [RNA-polymerase]-subunit kinase / negative regulation of protein localization to chromatin / transcription elongation-coupled chromatin remodeling / cellular response to cytokine stimulus / replication fork processing / Pausing and recovery of Tat-mediated HIV elongation / Tat-mediated HIV elongation arrest and recovery / RNA polymerase II transcribes snRNA genes / HIV elongation arrest and recovery / Pausing and recovery of HIV elongation / cyclin-dependent kinase / cyclin-dependent protein serine/threonine kinase activity / Tat-mediated elongation of the HIV-1 transcript / Formation of HIV-1 elongation complex containing HIV-1 Tat / Formation of HIV elongation complex in the absence of HIV Tat / regulation of DNA repair / RNA Polymerase II Transcription Elongation / Formation of RNA Pol II elongation complex / RNA Polymerase II Pre-transcription Events / RNA polymerase II CTD heptapeptide repeat kinase activity / transcription elongation factor complex / TP53 Regulates Transcription of DNA Repair Genes / transcription initiation at RNA polymerase II promoter / transcription elongation by RNA polymerase II / positive regulation of transcription elongation by RNA polymerase II / SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription / molecular condensate scaffold activity / PML body / transcription coactivator binding / cytoplasmic ribonucleoprotein granule / kinase activity / DNA-binding transcription factor binding / Estrogen-dependent gene expression / transcription by RNA polymerase II / cell population proliferation / protein phosphorylation / protein kinase activity / transcription cis-regulatory region binding / regulation of cell cycle / RNA polymerase II cis-regulatory region sequence-specific DNA binding / response to xenobiotic stimulus / cell division / protein serine kinase activity / DNA repair / protein serine/threonine kinase activity / chromatin binding / protein kinase binding / positive regulation of transcription by RNA polymerase II / DNA binding / nucleoplasm / ATP binding / nucleus / membrane / cytosol Similarity search - Function | ||||||
| Biological species |  HOMO SAPIENS (human) HOMO SAPIENS (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / OTHER / Resolution: 2.958 Å SYNCHROTRON / OTHER / Resolution: 2.958 Å | ||||||
 Authors Authors | Hole, A.J. / Baumli, S. / Wang, S. / Endicott, J.A. / Noble, M.E.M. | ||||||
 Citation Citation |  Journal: J.Med.Chem. / Year: 2013 Journal: J.Med.Chem. / Year: 2013Title: Comparative Structural and Functional Studies of 4-(Thiazol- 5-Yl)-2-(Phenylamino)Pyrimidine-5-Carbonitrile Cdk9 Inhibitors Suggest the Basis for Isotype Selectivity. Authors: Hole, A.J. / Baumli, S. / Shao, H. / Shi, S. / Pepper, C. / Fischer, P.M. / Wang, S. / Endicott, J.A. / Noble, M.E.M. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  4bch.cif.gz 4bch.cif.gz | 244.9 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb4bch.ent.gz pdb4bch.ent.gz | 199.5 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  4bch.json.gz 4bch.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  4bch_validation.pdf.gz 4bch_validation.pdf.gz | 815.8 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  4bch_full_validation.pdf.gz 4bch_full_validation.pdf.gz | 832.9 KB | Display | |
| Data in XML |  4bch_validation.xml.gz 4bch_validation.xml.gz | 27.1 KB | Display | |
| Data in CIF |  4bch_validation.cif.gz 4bch_validation.cif.gz | 34.4 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/bc/4bch https://data.pdbj.org/pub/pdb/validation_reports/bc/4bch ftp://data.pdbj.org/pub/pdb/validation_reports/bc/4bch ftp://data.pdbj.org/pub/pdb/validation_reports/bc/4bch | HTTPS FTP |
-Related structure data
| Related structure data |  4bcfC  4bciC  4bcjC  4bckC  4bcmC  4bcnC  4bcoC  4bcqC C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 38054.082 Da / Num. of mol.: 1 / Fragment: RESIDUES 2-330 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  HOMO SAPIENS (human) / Plasmid: PVL1392 / Production host: HOMO SAPIENS (human) / Plasmid: PVL1392 / Production host:  References: UniProt: P50750, cyclin-dependent kinase, [RNA-polymerase]-subunit kinase | ||||
|---|---|---|---|---|---|
| #2: Protein | Mass: 30119.426 Da / Num. of mol.: 1 / Fragment: RESIDUES 2-259 / Mutation: YES Source method: isolated from a genetically manipulated source Source: (gene. exp.)  HOMO SAPIENS (human) / Plasmid: PVL1392 / Production host: HOMO SAPIENS (human) / Plasmid: PVL1392 / Production host:  | ||||
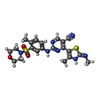
| #3: Chemical | ChemComp-T7Z / | ||||
| #4: Chemical | | #5: Water | ChemComp-HOH / | Has protein modification | Y | |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.3 Å3/Da / Density % sol: 46 % / Description: NONE |
|---|---|
| Crystal grow | Temperature: 277 K / pH: 6.2 Details: CRYSTALS WERE GROWN AT 4C USING 10-16% PEG 1000, 100MM NAK-PHOSPHATE PH 6.2, 500MM NACL, 4MM TCEP AS THE PRECIPITANT SOLUTION. THEY WERE SUBSEQUENTLY SOAKED IN THE PRESENCE OF COMPOUND. |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  Diamond Diamond  / Beamline: I03 / Wavelength: 0.9763 / Beamline: I03 / Wavelength: 0.9763 |
| Detector | Type: ADSC CCD / Detector: CCD |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.9763 Å / Relative weight: 1 |
| Reflection | Resolution: 2.96→87.09 Å / Num. obs: 23220 / % possible obs: 99.9 % / Redundancy: 5.24 % / Rmerge(I) obs: 0.07 / Net I/σ(I): 8.8 |
| Reflection shell | Resolution: 2.96→3.12 Å / Redundancy: 5.39 % / Rmerge(I) obs: 0.58 / Mean I/σ(I) obs: 1.33 / % possible all: 99.9 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure: OTHER / Resolution: 2.958→87.092 Å / Cor.coef. Fo:Fc: 0.955 / Cor.coef. Fo:Fc free: 0.939 / SU B: 23.924 / SU ML: 0.206 / Cross valid method: THROUGHOUT / ESU R: 0.57 / ESU R Free: 0.289 / Stereochemistry target values: MAXIMUM LIKELIHOOD / Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS.
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / VDW probe radii: 1.2 Å / Solvent model: MASK BULK SOLVENT | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 84.875 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.958→87.092 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller















 PDBj
PDBj