+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 3uit | ||||||
|---|---|---|---|---|---|---|---|
| Title | Overall structure of Patj/Pals1/Mals complex | ||||||
 Components Components | InaD-like protein, MAGUK p55 subfamily member 5, Protein lin-7 homolog B | ||||||
 Keywords Keywords | CELL ADHESION / L27 domain / CELL POLARIZATION | ||||||
| Function / homology |  Function and homology information Function and homology informationRHO GTPases Activate Rhotekin and Rhophilins / protein localization to myelin sheath abaxonal region / SARS-CoV-1 targets PDZ proteins in cell-cell junction / subapical complex / MPP7-DLG1-LIN7 complex / myelin assembly / establishment or maintenance of polarity of embryonic epithelium / regulation of synaptic assembly at neuromuscular junction / morphogenesis of an epithelial sheet / tight junction assembly ...RHO GTPases Activate Rhotekin and Rhophilins / protein localization to myelin sheath abaxonal region / SARS-CoV-1 targets PDZ proteins in cell-cell junction / subapical complex / MPP7-DLG1-LIN7 complex / myelin assembly / establishment or maintenance of polarity of embryonic epithelium / regulation of synaptic assembly at neuromuscular junction / morphogenesis of an epithelial sheet / tight junction assembly / Tight junction interactions / myelin sheath adaxonal region / SARS-CoV-2 targets PDZ proteins in cell-cell junction / regulation of transforming growth factor beta receptor signaling pathway / establishment of apical/basal cell polarity / Dopamine Neurotransmitter Release Cycle / microtubule organizing center organization / lateral loop / Schmidt-Lanterman incisure / establishment or maintenance of epithelial cell apical/basal polarity / peripheral nervous system myelin maintenance / cell-cell junction assembly / neurotransmitter secretion / tight junction / generation of neurons / apical junction complex / synaptic vesicle transport / central nervous system neuron development / exocytosis / positive regulation of epithelial cell migration / bicellular tight junction / postsynaptic density, intracellular component / endoplasmic reticulum-Golgi intermediate compartment membrane / regulation of microtubule cytoskeleton organization / protein localization to plasma membrane / PDZ domain binding / adherens junction / cerebral cortex development / postsynaptic density membrane / apical part of cell / cell-cell junction / protein transport / regulation of protein localization / presynapse / gene expression / basolateral plasma membrane / protein-macromolecule adaptor activity / perikaryon / apical plasma membrane / protein domain specific binding / axon / synapse / protein kinase binding / perinuclear region of cytoplasm / Golgi apparatus / protein-containing complex / extracellular exosome / ATP binding / identical protein binding / plasma membrane / cytoplasm Similarity search - Function | ||||||
| Biological species |    Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  SAD / Resolution: 2.05 Å SAD / Resolution: 2.05 Å | ||||||
 Authors Authors | Zhang, J. / Yang, X. / Long, J. / Shen, Y. | ||||||
 Citation Citation |  Journal: J.Biol.Chem. / Year: 2012 Journal: J.Biol.Chem. / Year: 2012Title: Structure of an L27 domain heterotrimer from cell polarity complex Patj/Pals1/Mals2 reveals mutually independent L27 domain assembly mode Authors: Zhang, J. / Yang, X. / Wang, Z. / Zhou, H. / Xie, X. / Shen, Y. / Long, J. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  3uit.cif.gz 3uit.cif.gz | 448.9 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb3uit.ent.gz pdb3uit.ent.gz | 370.8 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  3uit.json.gz 3uit.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ui/3uit https://data.pdbj.org/pub/pdb/validation_reports/ui/3uit ftp://data.pdbj.org/pub/pdb/validation_reports/ui/3uit ftp://data.pdbj.org/pub/pdb/validation_reports/ui/3uit | HTTPS FTP |
|---|
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 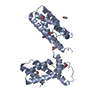
| |||||||||||||||
| 2 | 
| |||||||||||||||
| 3 | 
| |||||||||||||||
| 4 | 
| |||||||||||||||
| Unit cell |
| |||||||||||||||
| Components on special symmetry positions |
|
- Components
Components
| #1: Protein | Mass: 29856.209 Da / Num. of mol.: 4 Source method: isolated from a genetically manipulated source Details: Chimera of L27 domains of InaD-like protein (UNP residues 1-68) from Mouse, LINKER (LEVLFQGP), MAGUK p55 subfamily member 5 (UNP residues 119-232) from Human, LINKER (GGGLEVFQGP), Protein ...Details: Chimera of L27 domains of InaD-like protein (UNP residues 1-68) from Mouse, LINKER (LEVLFQGP), MAGUK p55 subfamily member 5 (UNP residues 119-232) from Human, LINKER (GGGLEVFQGP), Protein lin-7 homolog B (UNP residues 3-66) from Rat Source: (gene. exp.)    Homo sapiens (human) Homo sapiens (human)Production host:  References: UniProt: Q63ZW7, UniProt: Q8N3R9, UniProt: Q9Z252, UniProt: F1MAD2*PLUS #2: Chemical | ChemComp-ACT / #3: Water | ChemComp-HOH / | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 2 X-RAY DIFFRACTION / Number of used crystals: 2 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.58 Å3/Da / Density % sol: 52.33 % |
|---|---|
| Crystal grow | Temperature: 293 K / Method: vapor diffusion, sitting drop / pH: 7 Details: 2.8M sodium acetate, pH 7.0, VAPOR DIFFUSION, SITTING DROP, temperature 293K |
-Data collection
| Diffraction |
| ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Diffraction source |
| ||||||||||||||||||
| Detector |
| ||||||||||||||||||
| Radiation |
| ||||||||||||||||||
| Radiation wavelength | Wavelength: 0.9791 Å / Relative weight: 1 | ||||||||||||||||||
| Reflection | Resolution: 2.05→50 Å / Num. obs: 79261 / % possible obs: 87.7 % / Observed criterion σ(F): 2 / Observed criterion σ(I): 2 / Redundancy: 20.2 % / Biso Wilson estimate: 29.21 Å2 / Rmerge(I) obs: 0.063 / Rsym value: 0.081 / Net I/σ(I): 36.2 | ||||||||||||||||||
| Reflection shell | Resolution: 2.05→2.12 Å / % possible all: 100 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  SAD / Resolution: 2.05→38.863 Å / Occupancy max: 1 / Occupancy min: 1 / FOM work R set: 0.8479 / SU ML: 0.22 / σ(F): 2 / Phase error: 21.86 / Stereochemistry target values: ML SAD / Resolution: 2.05→38.863 Å / Occupancy max: 1 / Occupancy min: 1 / FOM work R set: 0.8479 / SU ML: 0.22 / σ(F): 2 / Phase error: 21.86 / Stereochemistry target values: ML
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å / Solvent model: FLAT BULK SOLVENT MODEL / Bsol: 49.246 Å2 / ksol: 0.343 e/Å3 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso max: 224.64 Å2 / Biso mean: 41.2893 Å2 / Biso min: 13.37 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.05→38.863 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Refine-ID: X-RAY DIFFRACTION / Total num. of bins used: 10
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Origin x: -43.3383 Å / Origin y: 27.7285 Å / Origin z: 34.3325 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group |
|
 Movie
Movie Controller
Controller












 PDBj
PDBj





