Entry Database : PDB / ID : 2be6Title 2.0 A crystal structure of the CaV1.2 IQ domain-Ca/CaM complex Calmodulin 2 Voltage-dependent L-type calcium channel alpha-1C subunit Keywords / / / / / / / / Function / homology Function Domain/homology Component
/ / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / Biological species Homo sapiens (human)Method / / / Resolution : 2 Å Authors Van Petegem, F. / Chatelain, F.C. / Minor Jr., D.L. Journal : Nat.Struct.Mol.Biol. / Year : 2005Title : Insights into voltage-gated calcium channel regulation from the structure of the Ca(V)1.2 IQ domain-Ca(2+)/calmodulin complexAuthors : van Petegem, F. / Chatelain, F.C. / Minor Jr., D.L. History Deposition Oct 23, 2005 Deposition site / Processing site Revision 1.0 Nov 15, 2005 Provider / Type Revision 1.1 May 1, 2008 Group Revision 1.2 Jul 13, 2011 Group / Version format complianceRevision 1.3 Oct 18, 2017 Group / Category Revision 1.4 Dec 21, 2022 Group / Derived calculationsCategory database_2 / pdbx_struct_conn_angle ... database_2 / pdbx_struct_conn_angle / struct_conn / struct_ref_seq_dif / struct_site Item _database_2.pdbx_DOI / _database_2.pdbx_database_accession ... _database_2.pdbx_DOI / _database_2.pdbx_database_accession / _pdbx_struct_conn_angle.ptnr1_auth_comp_id / _pdbx_struct_conn_angle.ptnr1_auth_seq_id / _pdbx_struct_conn_angle.ptnr1_label_alt_id / _pdbx_struct_conn_angle.ptnr1_label_asym_id / _pdbx_struct_conn_angle.ptnr1_label_atom_id / _pdbx_struct_conn_angle.ptnr1_label_comp_id / _pdbx_struct_conn_angle.ptnr1_label_seq_id / _pdbx_struct_conn_angle.ptnr2_auth_comp_id / _pdbx_struct_conn_angle.ptnr2_auth_seq_id / _pdbx_struct_conn_angle.ptnr2_label_asym_id / _pdbx_struct_conn_angle.ptnr2_label_atom_id / _pdbx_struct_conn_angle.ptnr2_label_comp_id / _pdbx_struct_conn_angle.ptnr3_auth_comp_id / _pdbx_struct_conn_angle.ptnr3_auth_seq_id / _pdbx_struct_conn_angle.ptnr3_label_alt_id / _pdbx_struct_conn_angle.ptnr3_label_asym_id / _pdbx_struct_conn_angle.ptnr3_label_atom_id / _pdbx_struct_conn_angle.ptnr3_label_comp_id / _pdbx_struct_conn_angle.ptnr3_label_seq_id / _pdbx_struct_conn_angle.value / _struct_conn.pdbx_dist_value / _struct_conn.pdbx_ptnr1_label_alt_id / _struct_conn.pdbx_ptnr2_label_alt_id / _struct_conn.ptnr1_auth_asym_id / _struct_conn.ptnr1_auth_comp_id / _struct_conn.ptnr1_auth_seq_id / _struct_conn.ptnr1_label_asym_id / _struct_conn.ptnr1_label_atom_id / _struct_conn.ptnr1_label_comp_id / _struct_conn.ptnr1_label_seq_id / _struct_conn.ptnr2_auth_asym_id / _struct_conn.ptnr2_auth_comp_id / _struct_conn.ptnr2_auth_seq_id / _struct_conn.ptnr2_label_asym_id / _struct_conn.ptnr2_label_atom_id / _struct_conn.ptnr2_label_comp_id / _struct_conn.ptnr2_label_seq_id / _struct_ref_seq_dif.details / _struct_site.pdbx_auth_asym_id / _struct_site.pdbx_auth_comp_id / _struct_site.pdbx_auth_seq_id Revision 1.5 May 22, 2024 Group / Category / chem_comp_bond
Show all Show less Remark 600 HETEROGEN Calcium 501, 502, 503, 504 are associated with chain A, Ca 506, 507, 508, 509 with chain ... HETEROGEN Calcium 501, 502, 503, 504 are associated with chain A, Ca 506, 507, 508, 509 with chain B and Ca 511, 512, 513, 514 with chain C. Ni 505 is associated with chain A, 510 with chain B and 515 with chain C.
 Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information Homo sapiens (human)
Homo sapiens (human) X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON /
SYNCHROTRON /  MAD / Resolution: 2 Å
MAD / Resolution: 2 Å  Authors
Authors Citation
Citation Journal: Nat.Struct.Mol.Biol. / Year: 2005
Journal: Nat.Struct.Mol.Biol. / Year: 2005 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 2be6.cif.gz
2be6.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb2be6.ent.gz
pdb2be6.ent.gz PDB format
PDB format 2be6.json.gz
2be6.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/be/2be6
https://data.pdbj.org/pub/pdb/validation_reports/be/2be6 ftp://data.pdbj.org/pub/pdb/validation_reports/be/2be6
ftp://data.pdbj.org/pub/pdb/validation_reports/be/2be6 Links
Links Assembly
Assembly
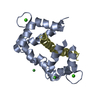



 Components
Components Homo sapiens (human) / Gene: CALM2 / Plasmid: pEGST / Production host:
Homo sapiens (human) / Gene: CALM2 / Plasmid: pEGST / Production host: 
 Homo sapiens (human) / Gene: CACNA1C, CACH2, CACN2, CACNL1A1, CCHL1A1 / Plasmid: pEGST / Production host:
Homo sapiens (human) / Gene: CACNA1C, CACH2, CACN2, CACNL1A1, CCHL1A1 / Plasmid: pEGST / Production host: 
 X-RAY DIFFRACTION / Number of used crystals: 2
X-RAY DIFFRACTION / Number of used crystals: 2  Sample preparation
Sample preparation Processing
Processing MAD / Resolution: 2→20 Å / Cor.coef. Fo:Fc: 0.95 / Cor.coef. Fo:Fc free: 0.923 / SU B: 8.165 / SU ML: 0.13 / TLS residual ADP flag: LIKELY RESIDUAL / Cross valid method: THROUGHOUT / σ(F): 0 / ESU R: 0.2 / ESU R Free: 0.182 / Stereochemistry target values: MAXIMUM LIKELIHOOD
MAD / Resolution: 2→20 Å / Cor.coef. Fo:Fc: 0.95 / Cor.coef. Fo:Fc free: 0.923 / SU B: 8.165 / SU ML: 0.13 / TLS residual ADP flag: LIKELY RESIDUAL / Cross valid method: THROUGHOUT / σ(F): 0 / ESU R: 0.2 / ESU R Free: 0.182 / Stereochemistry target values: MAXIMUM LIKELIHOOD Movie
Movie Controller
Controller













 PDBj
PDBj


































