[English] 日本語
 Yorodumi
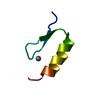
Yorodumi- PDB-2omm: GNNQQNY peptide corresponding to residues 7-13 of yeast prion sup35 -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 2omm | ||||||
|---|---|---|---|---|---|---|---|
| Title | GNNQQNY peptide corresponding to residues 7-13 of yeast prion sup35 | ||||||
 Components Components | GNNQQNY peptide corresponding to residues 7-13 of yeast prion sup35 | ||||||
 Keywords Keywords | PROTEIN FIBRIL / steric zipper / glutamine zipper / polar zipper / asparagine zipper | ||||||
| Function / homology |  Function and homology information Function and homology informationEukaryotic Translation Termination / translation release factor complex / translation release factor activity / nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay / Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) / Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) / translational termination / cytoplasmic stress granule / regulation of translation / ribosome binding ...Eukaryotic Translation Termination / translation release factor complex / translation release factor activity / nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay / Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) / Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) / translational termination / cytoplasmic stress granule / regulation of translation / ribosome binding / Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement / translation / GTPase activity / mRNA binding / GTP binding / identical protein binding / cytoplasm / cytosol Similarity search - Function | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2 Å MOLECULAR REPLACEMENT / Resolution: 2 Å | ||||||
 Authors Authors | Sawaya, M.R. / Nelson, R. / Eisenberg, D. | ||||||
 Citation Citation |  Journal: Nature / Year: 2007 Journal: Nature / Year: 2007Title: Atomic structures of amyloid cross-beta spines reveal varied steric zippers. Authors: Sawaya, M.R. / Sambashivan, S. / Nelson, R. / Ivanova, M.I. / Sievers, S.A. / Apostol, M.I. / Thompson, M.J. / Balbirnie, M. / Wiltzius, J.J. / McFarlane, H.T. / Madsen, A.O. / Riekel, C. / Eisenberg, D. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  2omm.cif.gz 2omm.cif.gz | 8.6 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb2omm.ent.gz pdb2omm.ent.gz | 5.4 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  2omm.json.gz 2omm.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/om/2omm https://data.pdbj.org/pub/pdb/validation_reports/om/2omm ftp://data.pdbj.org/pub/pdb/validation_reports/om/2omm ftp://data.pdbj.org/pub/pdb/validation_reports/om/2omm | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  2okzC  2ol9C  2olxC  2ompC  2omqC  2on9C  2onaC  2onvC  2onwC  2onxC  1yjpS S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| Unit cell |
| ||||||||
| Details | One sheet of the steric zipper can be generated by repeated application of the crystallographic unit cell translation along the b axis. The second sheet of the steric zipper can be generated by application of the crystallographic operator -X+1,-1/2+Y,1/2-Z, and repeated unit cell translations of this strand along the b axis. |
- Components
Components
| #1: Protein/peptide | Mass: 836.807 Da / Num. of mol.: 1 / Fragment: residues 7-13 / Source method: obtained synthetically / References: UniProt: P05453*PLUS |
|---|---|
| #2: Water | ChemComp-HOH / |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal grow | Temperature: 298 K / Method: evaporation, recrystallization Details: peptide was dissolved at 10 mg/mL in water, quickly filtered, and left to sit at room temperature, EVAPORATION, RECRYSTALLIZATION, temperature 298K |
|---|
-Data collection
| Diffraction | Mean temperature: 100 K | |||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  ESRF ESRF  / Beamline: ID13 / Wavelength: 0.9466 / Beamline: ID13 / Wavelength: 0.9466 | |||||||||||||||||||||||||||||||||||
| Detector | Type: MAR CCD 165 mm / Detector: CCD / Date: Jul 16, 2005 | |||||||||||||||||||||||||||||||||||
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray | |||||||||||||||||||||||||||||||||||
| Radiation wavelength | Wavelength: 0.9466 Å / Relative weight: 1 | |||||||||||||||||||||||||||||||||||
| Reflection | Resolution: 2→19.81 Å / Num. obs: 380 / % possible obs: 96.4 % / Observed criterion σ(I): -3 / Biso Wilson estimate: 2.446 Å2 / Rmerge(I) obs: 0.248 / Net I/σ(I): 4.61 | |||||||||||||||||||||||||||||||||||
| Reflection shell |
|
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: pdb entry 1yjp Resolution: 2→19.81 Å / Cor.coef. Fo:Fc: 0.904 / Cor.coef. Fo:Fc free: 0.885 / SU B: 5.08 / SU ML: 0.141 / Cross valid method: THROUGHOUT / σ(F): 0 / ESU R: 0.382 / ESU R Free: 0.218 / Stereochemistry target values: MAXIMUM LIKELIHOOD / Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.4 Å / Solvent model: MASK | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 12.533 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2→19.81 Å /
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 2→2.053 Å / Total num. of bins used: 20
|
 Movie
Movie Controller
Controller








 PDBj
PDBj







