[English] 日本語
 Yorodumi
Yorodumi- PDB-2onw: Structure of SSTSSA, a fibril forming peptide from Bovine Pancrea... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 2onw | ||||||
|---|---|---|---|---|---|---|---|
| Title | Structure of SSTSSA, a fibril forming peptide from Bovine Pancreatic Ribonuclease (RNase A, residues 15-20) | ||||||
 Components Components | fibril forming peptide from Bovine Pancreatic Ribonuclease (RNase A) | ||||||
 Keywords Keywords | PROTEIN FIBRIL / parallel face-to-face-Up/Up beta sheets / steric zipper | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.51 Å MOLECULAR REPLACEMENT / Resolution: 1.51 Å | ||||||
 Authors Authors | Sambashivan, S. / Sawaya, M.R. / Eisenberg, D. | ||||||
 Citation Citation |  Journal: Nature / Year: 2007 Journal: Nature / Year: 2007Title: Atomic structures of amyloid cross-beta spines reveal varied steric zippers. Authors: Sawaya, M.R. / Sambashivan, S. / Nelson, R. / Ivanova, M.I. / Sievers, S.A. / Apostol, M.I. / Thompson, M.J. / Balbirnie, M. / Wiltzius, J.J. / McFarlane, H.T. / Madsen, A.O. / Riekel, C. / Eisenberg, D. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  2onw.cif.gz 2onw.cif.gz | 9.9 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb2onw.ent.gz pdb2onw.ent.gz | 6.9 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  2onw.json.gz 2onw.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/on/2onw https://data.pdbj.org/pub/pdb/validation_reports/on/2onw ftp://data.pdbj.org/pub/pdb/validation_reports/on/2onw ftp://data.pdbj.org/pub/pdb/validation_reports/on/2onw | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  2okzC  2ol9C  2olxC  2ommC  2ompC  2omqC  2on9C  2onaC  2onvC  2onxC C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 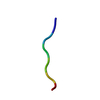
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| Unit cell |
| ||||||||
| Details | The biological unit is a pair of sheets. Each sheet is comprised of beta strands generated by unit cell translations along the y-axis. The second sheet in the pair-of-sheet structures is generated by applying the operator -x+1, y+1/2, -z+1/2. In the second sheet also, beta strands are generated by unit cell translations along the y-axis. |
- Components
Components
| #1: Protein/peptide | Mass: 522.508 Da / Num. of mol.: 1 / Fragment: hinge loop region (residues 15-20) / Source method: obtained synthetically / Details: The peptide SSTSAA was commercially synthesized. |
|---|---|
| #2: Water | ChemComp-HOH / |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal grow | Temperature: 291 K / Method: vapor diffusion, hanging drop / pH: 7.5 Details: Peptide concentration: 30 mg/ml, Reservoir: 0.1M HEPES-Na, pH 7.5, 10% v/v isopropanol, 20% PEG4000, VAPOR DIFFUSION, HANGING DROP, temperature 291K |
|---|
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  ESRF ESRF  / Beamline: ID13 / Wavelength: 0.9466 / Beamline: ID13 / Wavelength: 0.9466 |
| Detector | Type: MARRESEARCH / Detector: CCD / Date: Jan 1, 2005 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.9466 Å / Relative weight: 1 |
| Reflection | Resolution: 1.5→80 Å / Num. obs: 474 / % possible obs: 89.3 % / Biso Wilson estimate: 6.599 Å2 / Rmerge(I) obs: 0.174 / Χ2: 1.067 / Net I/σ(I): 3.9 |
| Reflection shell | Resolution: 1.5→1.62 Å / Rmerge(I) obs: 0.434 / Num. unique all: 98 / Χ2: 1.084 / % possible all: 93.3 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 5 residue beta strand SSTSA with the C-terminal alanine absent Resolution: 1.51→21 Å / Cor.coef. Fo:Fc: 0.973 / Cor.coef. Fo:Fc free: 0.903 / SU B: 2.481 / SU ML: 0.04 / Cross valid method: THROUGHOUT / σ(F): 0 / ESU R: 0.151 / ESU R Free: 0.102 / Stereochemistry target values: MAXIMUM LIKELIHOOD / Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.4 Å / Solvent model: MASK | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 5.291 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.51→21 Å /
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 1.51→1.551 Å / Total num. of bins used: 20
|
 Movie
Movie Controller
Controller







 PDBj
PDBj
