[English] 日本語
 Yorodumi
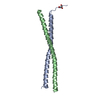
Yorodumi- PDB-2n9b: Solution NMR Structure of Antiparallel Myosin-10:GCN4 Tandem Coil... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 2n9b | ||||||
|---|---|---|---|---|---|---|---|
| Title | Solution NMR Structure of Antiparallel Myosin-10:GCN4 Tandem Coiled-Coil | ||||||
 Components Components | Unconventional myosin-X, General control protein GCN4 fusion | ||||||
 Keywords Keywords | MOTOR PROTEIN/TRANSCRIPTION / anti-parallel coiled-coil / coiled-coil / MOTOR PROTEIN-TRANSCRIPTION complex | ||||||
| Function / homology |  Function and homology information Function and homology informationplus-end directed microfilament motor activity / cytoskeleton-dependent intracellular transport / filopodium tip / FCERI mediated MAPK activation / protein localization to nuclear periphery / Activation of the AP-1 family of transcription factors / regulation of filopodium assembly / response to amino acid starvation / negative regulation of ribosomal protein gene transcription by RNA polymerase II / positive regulation of cellular response to amino acid starvation ...plus-end directed microfilament motor activity / cytoskeleton-dependent intracellular transport / filopodium tip / FCERI mediated MAPK activation / protein localization to nuclear periphery / Activation of the AP-1 family of transcription factors / regulation of filopodium assembly / response to amino acid starvation / negative regulation of ribosomal protein gene transcription by RNA polymerase II / positive regulation of cellular response to amino acid starvation / mediator complex binding / Oxidative Stress Induced Senescence / filopodium membrane / myosin complex / microfilament motor activity / phosphatidylinositol-3,4,5-trisphosphate binding / TFIID-class transcription factor complex binding / amino acid biosynthetic process / positive regulation of RNA polymerase II transcription preinitiation complex assembly / positive regulation of transcription initiation by RNA polymerase II / cellular response to nutrient levels / ruffle / cellular response to amino acid starvation / filopodium / RNA polymerase II transcription regulator complex / actin filament binding / regulation of cell shape / lamellipodium / DNA-binding transcription activator activity, RNA polymerase II-specific / cell cortex / transcription regulator complex / sequence-specific DNA binding / RNA polymerase II-specific DNA-binding transcription factor binding / DNA-binding transcription factor activity, RNA polymerase II-specific / calmodulin binding / intracellular signal transduction / RNA polymerase II cis-regulatory region sequence-specific DNA binding / DNA-binding transcription factor activity / chromatin binding / negative regulation of transcription by RNA polymerase II / signal transduction / positive regulation of transcription by RNA polymerase II / ATP binding / identical protein binding / nucleus / cytosol Similarity search - Function | ||||||
| Biological species |   | ||||||
| Method | SOLUTION NMR / simulated annealing | ||||||
| Model details | lowest energy, model1 | ||||||
 Authors Authors | Vavra, K.C. / Xia, Y. / Rock, R.S. | ||||||
 Citation Citation |  Journal: Biophys.J. / Year: 2016 Journal: Biophys.J. / Year: 2016Title: Competition between Coiled-Coil Structures and the Impact on Myosin-10 Bundle Selection Authors: Vavra, K.C. / Xia, Y. / Rock, R.S. #1:  Journal: Proc.Natl.Acad.Sci.USA / Year: 2012 Journal: Proc.Natl.Acad.Sci.USA / Year: 2012Title: Antiparallel coiled-coil-mediated dimerization of myosin X. Authors: Lu, Q. / Ye, F. / Wei, Z. / Wen, Z. / Zhang, M. #2:  Journal: Science / Year: 1991 Journal: Science / Year: 1991Title: X-ray structure of the GCN4 leucine zipper, a two-stranded, parallel coiled coil Authors: Klemm, J.D. / Kim, P.S. / Alber, T. #3: Journal: Proc.Natl.Acad.Sci.USA / Year: 2008 Title: A myosin motor that selects bundled actin for motility Authors: Nagy, S. / Ricca, B.L. / Norstrom, M.F. / Courson, D.F. / Brawley, C.M. / Smithback, P.A. / Rock, R.S. #4: Journal: Proc.Natl.Acad.Sci.USA / Year: 1997 Title: Spare the rod, spoil the regulation: necessity for a myosin rod Authors: Trybus, K.M. / Freyzon, Y. / Faust, L.Z. / Sweeney, H. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  2n9b.cif.gz 2n9b.cif.gz | 456.2 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb2n9b.ent.gz pdb2n9b.ent.gz | 386.1 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  2n9b.json.gz 2n9b.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  2n9b_validation.pdf.gz 2n9b_validation.pdf.gz | 536.6 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  2n9b_full_validation.pdf.gz 2n9b_full_validation.pdf.gz | 724.6 KB | Display | |
| Data in XML |  2n9b_validation.xml.gz 2n9b_validation.xml.gz | 36 KB | Display | |
| Data in CIF |  2n9b_validation.cif.gz 2n9b_validation.cif.gz | 48.8 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/n9/2n9b https://data.pdbj.org/pub/pdb/validation_reports/n9/2n9b ftp://data.pdbj.org/pub/pdb/validation_reports/n9/2n9b ftp://data.pdbj.org/pub/pdb/validation_reports/n9/2n9b | HTTPS FTP |
-Related structure data
| Related structure data | |
|---|---|
| Similar structure data | |
| Other databases |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: Protein | Mass: 8200.290 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Gene: MYO10, GCN4, AAS3, ARG9, YEL009C / Strain: ATCC 204508 / S288c / Production host:  |
|---|
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR Details: Tandem antiparallel coiled-coil fusion derived from Bos taurus myosin-10 coiled-coil and GCN4-p1. Structure solved by solution NMR. Tertiary structure supported by Small-Angle X-ray scattering (see reference). | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
|
- Sample preparation
Sample preparation
| Details | Contents: 1.1 mM [U-100% 13C; U-100% 15N] entity-1, 100 mM potassium phosphate-2, 1 mM EDTA-3, 0.03 % sodium azide-4, 90% H2O/10% D2O Solvent system: 90% H2O/10% D2O | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample |
| ||||||||||||||||||||
| Sample conditions | Ionic strength: 0.6 / pH: 6.5 / Pressure: ambient / Temperature: 308 K |
-NMR measurement
| NMR spectrometer |
|
|---|
- Processing
Processing
| NMR software |
| ||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method: simulated annealing / Software ordinal: 1 | ||||||||||||||||||||||||||||
| NMR constraints | NOE constraints total: 1561 / NOE intraresidue total count: 332 / NOE long range total count: 149 / NOE medium range total count: 610 / NOE sequential total count: 470 / Hydrogen bond constraints total count: 294 / Protein phi angle constraints total count: 147 / Protein psi angle constraints total count: 147 | ||||||||||||||||||||||||||||
| NMR representative | Selection criteria: lowest energy | ||||||||||||||||||||||||||||
| NMR ensemble | Conformer selection criteria: structures with the lowest energy Conformers calculated total number: 100 / Conformers submitted total number: 10 |
 Movie
Movie Controller
Controller













 PDBj
PDBj









 HSQC
HSQC