[English] 日本語
 Yorodumi
Yorodumi- PDB-2bda: Porcine pancreatic elastase complexed with N-acetyl-NPI and Ala-A... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 2bda | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Porcine pancreatic elastase complexed with N-acetyl-NPI and Ala-Ala at pH 5.0 | |||||||||
 Components Components |
| |||||||||
 Keywords Keywords | HYDROLASE / SERINE PROTEINASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationpancreatic elastase / serine-type endopeptidase activity / proteolysis / extracellular space / metal ion binding Similarity search - Function | |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.8 Å MOLECULAR REPLACEMENT / Resolution: 1.8 Å | |||||||||
 Authors Authors | Liu, B. / Schofield, C.J. / Wilmouth, R.C. | |||||||||
 Citation Citation |  Journal: J.Biol.Chem. / Year: 2006 Journal: J.Biol.Chem. / Year: 2006Title: Structural analyses on intermediates in serine protease catalysis Authors: Liu, B. / Schofield, C.J. / Wilmouth, R.C. #1:  Journal: Nat.Struct.Biol. / Year: 1997 Journal: Nat.Struct.Biol. / Year: 1997Title: Structure of a specific acyl-enzyme complex formed between beta-casomorphin-7 and porcine pancreatic elastase Authors: Wilmouth, R.C. / Clifton, I.J. / Robinson, C.V. / Roach, P.L. / Aplin, R.T. / Westwood, N.J. / Hajdu, J. / Schofield, C.J. #2:  Journal: Nat.Struct.Biol. / Year: 2001 Journal: Nat.Struct.Biol. / Year: 2001Title: X-ray snapshots of serine protease catalysis reveal a tetrahedral intermediate Authors: Wilmouth, R.C. / Edman, K. / Neutze, R. / Wright, P.A. / Clifton, I.J. / Schneider, T.R. / Schofield, C.J. / Hajdu, J. #3:  Journal: J.BIOL.CHEM. / Year: 2002 Journal: J.BIOL.CHEM. / Year: 2002Title: X-ray structure of a serine protease acyl-enzyme complex at 0.95-A resolution Authors: Katona, G. / Wilmouth, R.C. / Wright, P.A. / Berglund, G.I. / Hajdu, J. / Neutze, R. / Schofield, C.J. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  2bda.cif.gz 2bda.cif.gz | 65.3 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb2bda.ent.gz pdb2bda.ent.gz | 45.9 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  2bda.json.gz 2bda.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/bd/2bda https://data.pdbj.org/pub/pdb/validation_reports/bd/2bda ftp://data.pdbj.org/pub/pdb/validation_reports/bd/2bda ftp://data.pdbj.org/pub/pdb/validation_reports/bd/2bda | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  2bb4C  2bd2C  2bd3C  2bd4C  2bd5C  2bd7C  2bd8C  2bd9C  2bdbC  2bdcC  2h1uC  3estS  2bd6 C: citing same article ( S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 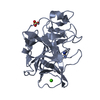
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein/peptide | Mass: 484.546 Da / Num. of mol.: 1 / Source method: obtained synthetically / Details: synthetic peptide / Source: (synth.)  Homo sapiens (human) Homo sapiens (human) | ||
|---|---|---|---|
| #2: Protein | Mass: 25928.031 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  | ||
| #3: Chemical | ChemComp-CA / | ||
| #4: Chemical | ChemComp-SO4 / | ||
| #5: Water | ChemComp-HOH / | ||
| Has protein modification | Y | ||
| Nonpolymer details | OG SER A 195, C ILE P 7 AND N ALA P 1001 ARE COVALENTLY| Sequence details | THIS CONFLICT IS BASED ON THE REFERENCE 3 IN SEQUENCE DATABASE, P00772 IN SWISS-PROT. THE PEPTIDE ...THIS CONFLICT IS BASED ON THE REFERENCE 3 IN SEQUENCE DATABASE, P00772 IN SWISS-PROT. THE PEPTIDE CHAIN P HAS ACETYLATIO | |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.04 Å3/Da / Density % sol: 39.85 % |
|---|---|
| Crystal grow | Temperature: 294 K / Method: vapor diffusion, hanging drop / pH: 5 Details: 25MM SODIUM SULPHATE, 25MM SODIUM ACETATE (PH 5.0), 25 MG/ML PPE AND 17.5 MG/ML AcNPI, AND THEN SOAKED WITH ALA-ALA (2 MG) IN 250MM SODIUM ACETATE (PH 5.0) (5 MICROLITRES) FOR 30 MIN, VAPOR ...Details: 25MM SODIUM SULPHATE, 25MM SODIUM ACETATE (PH 5.0), 25 MG/ML PPE AND 17.5 MG/ML AcNPI, AND THEN SOAKED WITH ALA-ALA (2 MG) IN 250MM SODIUM ACETATE (PH 5.0) (5 MICROLITRES) FOR 30 MIN, VAPOR DIFFUSION, HANGING DROP, temperature 294K |
-Data collection
| Diffraction | Mean temperature: 127 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  SLS SLS  / Beamline: X06SA / Wavelength: 0.98 Å / Beamline: X06SA / Wavelength: 0.98 Å |
| Detector | Type: MARRESEARCH / Detector: CCD / Date: May 2, 2005 / Details: mirrors |
| Radiation | Monochromator: Si(111) / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.98 Å / Relative weight: 1 |
| Reflection | Resolution: 1.8→19.6 Å / Num. obs: 20470 / % possible obs: 99.5 % / Observed criterion σ(F): 0 / Observed criterion σ(I): 0 / Redundancy: 3.2 % / Biso Wilson estimate: 14.9 Å2 / Rmerge(I) obs: 0.086 / Net I/σ(I): 11.1 |
| Reflection shell | Resolution: 1.8→1.9 Å / Redundancy: 3.3 % / Rmerge(I) obs: 0.46 / Mean I/σ(I) obs: 2.4 / Num. unique all: 2950 / % possible all: 100 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 3EST Resolution: 1.8→19.6 Å / Cor.coef. Fo:Fc: 0.957 / Cor.coef. Fo:Fc free: 0.935 / SU B: 5.434 / SU ML: 0.084 / TLS residual ADP flag: LIKELY RESIDUAL / Cross valid method: THROUGHOUT / σ(F): 0 / ESU R: 0.129 / ESU R Free: 0.124 / Stereochemistry target values: MAXIMUM LIKELIHOOD
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.2 Å / Solvent model: BABINET MODEL WITH MASK | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 17.773 Å2
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.8→19.6 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 1.8→1.847 Å / Total num. of bins used: 20
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Origin x: -4.272 Å / Origin y: 28.184 Å / Origin z: 42.571 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group | Selection: ALL |
 Movie
Movie Controller
Controller


















 PDBj
PDBj





