[English] 日本語
 Yorodumi
Yorodumi- PDB-1rml: NMR STUDY OF ACID FIBROBLAST GROWTH FACTOR BOUND TO 1,3,6-NAPHTHA... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1rml | ||||||
|---|---|---|---|---|---|---|---|
| Title | NMR STUDY OF ACID FIBROBLAST GROWTH FACTOR BOUND TO 1,3,6-NAPHTHALENE TRISULPHONATE, 26 STRUCTURES | ||||||
 Components Components | ACIDIC FIBROBLAST GROWTH FACTOR | ||||||
 Keywords Keywords | GROWTH FACTOR / FIBROBLAST GROWTH FACTOR / ANTITUMORAL / NAPHTHALENE | ||||||
| Function / homology |  Function and homology information Function and homology informationmesonephric epithelium development / branch elongation involved in ureteric bud branching / regulation of endothelial tube morphogenesis / FGFR3b ligand binding and activation / regulation of endothelial cell chemotaxis to fibroblast growth factor / Signaling by activated point mutants of FGFR3 / FGFR3c ligand binding and activation / Phospholipase C-mediated cascade; FGFR3 / FGFR2b ligand binding and activation / fibroblast growth factor receptor binding ...mesonephric epithelium development / branch elongation involved in ureteric bud branching / regulation of endothelial tube morphogenesis / FGFR3b ligand binding and activation / regulation of endothelial cell chemotaxis to fibroblast growth factor / Signaling by activated point mutants of FGFR3 / FGFR3c ligand binding and activation / Phospholipase C-mediated cascade; FGFR3 / FGFR2b ligand binding and activation / fibroblast growth factor receptor binding / FGFR2c ligand binding and activation / Activated point mutants of FGFR2 / Phospholipase C-mediated cascade; FGFR2 / FGFR4 ligand binding and activation / FGFR1b ligand binding and activation / Phospholipase C-mediated cascade; FGFR4 / Signaling by activated point mutants of FGFR1 / FGFR1c ligand binding and activation / organ induction / Downstream signaling of activated FGFR1 / Phospholipase C-mediated cascade: FGFR1 / S100 protein binding / Signaling by FGFR2 IIIa TM / PI-3K cascade:FGFR3 / PI-3K cascade:FGFR2 / PI-3K cascade:FGFR4 / PI-3K cascade:FGFR1 / positive regulation of sprouting angiogenesis / positive regulation of MAP kinase activity / positive regulation of intracellular signal transduction / positive regulation of cell division / PI3K Cascade / fibroblast growth factor receptor signaling pathway / activation of protein kinase B activity / anatomical structure morphogenesis / SHC-mediated cascade:FGFR3 / SHC-mediated cascade:FGFR2 / SHC-mediated cascade:FGFR4 / SHC-mediated cascade:FGFR1 / FRS-mediated FGFR3 signaling / FRS-mediated FGFR2 signaling / FRS-mediated FGFR4 signaling / Signaling by FGFR3 in disease / FRS-mediated FGFR1 signaling / Signaling by FGFR2 in disease / neurogenesis / Signaling by FGFR1 in disease / extracellular matrix / positive regulation of endothelial cell migration / lung development / regulation of cell migration / epithelial cell proliferation / positive regulation of epithelial cell proliferation / growth factor activity / Negative regulation of FGFR3 signaling / Negative regulation of FGFR2 signaling / Negative regulation of FGFR4 signaling / wound healing / Negative regulation of FGFR1 signaling / positive regulation of cholesterol biosynthetic process / integrin binding / positive regulation of angiogenesis / Constitutive Signaling by Aberrant PI3K in Cancer / PIP3 activates AKT signaling / heparin binding / cellular response to heat / PI5P, PP2A and IER3 Regulate PI3K/AKT Signaling / RAF/MAP kinase cascade / angiogenesis / cell cortex / positive regulation of ERK1 and ERK2 cascade / positive regulation of MAPK cascade / positive regulation of cell migration / positive regulation of cell population proliferation / signal transduction / positive regulation of transcription by RNA polymerase II / extracellular space / extracellular region / nucleoplasm / nucleus / cytosol / cytoplasm Similarity search - Function | ||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||
| Method | SOLUTION NMR / VARIABLE TARGET FUNCTION | ||||||
 Authors Authors | Lozano, R.M. / Jimenez, M.A. / Santoro, J. / Rico, M. / Gimenez-Gallego, G. | ||||||
 Citation Citation |  Journal: J.Mol.Biol. / Year: 1998 Journal: J.Mol.Biol. / Year: 1998Title: Solution structure of acidic fibroblast growth factor bound to 1,3, 6-naphthalenetrisulfonate: a minimal model for the anti-tumoral action of suramins and suradistas. Authors: Lozano, R.M. / Jimenez, M. / Santoro, J. / Rico, M. / Gimenez-Gallego, G. #1:  Journal: J.Mol.Biol. / Year: 1996 Journal: J.Mol.Biol. / Year: 1996Title: Three-Dimensional Structure of Acidic Fibroblast Growth Factor in Solution: Effects of Binding to a Heparin Functional Analog Authors: Pineda-Lucena, A. / Jimenez, M.A. / Lozano, R.M. / Nieto, J.L. / Santoro, J. / Rico, M. / Gimenez-Gallego, G. #2:  Journal: J.Mol.Biol. / Year: 1994 Journal: J.Mol.Biol. / Year: 1994Title: 1H-NMR Assignment and Solution Structure of Human Acidic Fibroblast Growth Factor Activated by Inositol Hexasulfate Authors: Pineda-Lucena, A. / Jimenez, M.A. / Nieto, J.L. / Santoro, J. / Rico, M. / Gimenez-Gallego, G. #3:  Journal: J.Mol.Biol. / Year: 1994 Journal: J.Mol.Biol. / Year: 1994Title: Erratum. 1H-NMR Assignment and Solution Structure of Human Acidic Fibroblast Growth Factor Activated by Inositol Hexasulfate Authors: Pineda-Lucena, A. / Jimenez, M.A. / Nieto, J.L. / Santoro, J. / Rico, M. / Gimenez-Gallego, G. | ||||||
| History |
|
- Structure visualization
Structure visualization
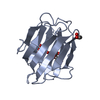
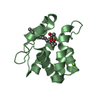
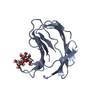
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1rml.cif.gz 1rml.cif.gz | 1 MB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1rml.ent.gz pdb1rml.ent.gz | 879.8 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1rml.json.gz 1rml.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  1rml_validation.pdf.gz 1rml_validation.pdf.gz | 447.7 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  1rml_full_validation.pdf.gz 1rml_full_validation.pdf.gz | 785.9 KB | Display | |
| Data in XML |  1rml_validation.xml.gz 1rml_validation.xml.gz | 122.1 KB | Display | |
| Data in CIF |  1rml_validation.cif.gz 1rml_validation.cif.gz | 146.9 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/rm/1rml https://data.pdbj.org/pub/pdb/validation_reports/rm/1rml ftp://data.pdbj.org/pub/pdb/validation_reports/rm/1rml ftp://data.pdbj.org/pub/pdb/validation_reports/rm/1rml | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: Protein | Mass: 17482.697 Da / Num. of mol.: 1 / Fragment: RESIDUES 23 - 154 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Plasmid: PMG47 / Production host: Homo sapiens (human) / Plasmid: PMG47 / Production host:  |
|---|---|
| #2: Chemical | ChemComp-NTS / |
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
|
- Sample preparation
Sample preparation
| Details | Contents: WATER |
|---|---|
| Sample conditions | pH: 6 / Temperature: 298 K |
| Crystal grow | *PLUS Method: other / Details: NMR |
-NMR measurement
| NMR spectrometer | Type: Bruker AMX 600 / Manufacturer: Bruker / Model: AMX 600 / Field strength: 600 MHz |
|---|
- Processing
Processing
| NMR software |
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method: VARIABLE TARGET FUNCTION / Software ordinal: 1 | |||||||||
| NMR ensemble | Conformers calculated total number: 26 / Conformers submitted total number: 26 |
 Movie
Movie Controller
Controller









 PDBj
PDBj