[English] 日本語
 Yorodumi
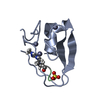
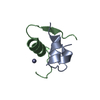
Yorodumi- PDB-1kde: NORTH-ATLANTIC OCEAN POUT ANTIFREEZE PROTEIN TYPE III ISOFORM HPL... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1kde | ||||||
|---|---|---|---|---|---|---|---|
| Title | NORTH-ATLANTIC OCEAN POUT ANTIFREEZE PROTEIN TYPE III ISOFORM HPLC12 MUTANT, NMR, 22 STRUCTURES | ||||||
 Components Components | ANTIFREEZE PROTEIN TYPE III ISOFORM HPLC12 MUTANT | ||||||
 Keywords Keywords | ANTIFREEZE PROTEIN / ICE BINDING PROTEIN / THERMAL HYSTERESIS PROTEIN / GLYCOPROTEIN | ||||||
| Function / homology |  Function and homology information Function and homology information | ||||||
| Biological species |  Macrozoarces americanus (ocean pout) Macrozoarces americanus (ocean pout) | ||||||
| Method | SOLUTION NMR / DG_SUB_EMBED, DGSA, REFINE | ||||||
 Authors Authors | Sonnichsen, F.D. / Deluca, C.I. / Davies, P.L. / Sykes, B.D. | ||||||
 Citation Citation |  Journal: Structure / Year: 1996 Journal: Structure / Year: 1996Title: Refined solution structure of type III antifreeze protein: hydrophobic groups may be involved in the energetics of the protein-ice interaction. Authors: Sonnichsen, F.D. / DeLuca, C.I. / Davies, P.L. / Sykes, B.D. #1:  Journal: Protein Sci. / Year: 1994 Journal: Protein Sci. / Year: 1994Title: Structure-Function Relationship in the Globular Type III Antifreeze Protein: Identification of a Cluster of Surface Residues Required for Binding to Ice Authors: Chao, H. / Sonnichsen, F.D. / Deluca, C.I. / Sykes, B.D. / Davies, P.L. #2:  Journal: Science / Year: 1993 Journal: Science / Year: 1993Title: The Nonhelical Structure of Antifreeze Protein Type III Authors: Sonnichsen, F.D. / Sykes, B.D. / Chao, H. / Davies, P.L. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1kde.cif.gz 1kde.cif.gz | 422.4 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1kde.ent.gz pdb1kde.ent.gz | 353.1 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1kde.json.gz 1kde.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/kd/1kde https://data.pdbj.org/pub/pdb/validation_reports/kd/1kde ftp://data.pdbj.org/pub/pdb/validation_reports/kd/1kde ftp://data.pdbj.org/pub/pdb/validation_reports/kd/1kde | HTTPS FTP |
|---|
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: Protein | Mass: 7525.890 Da / Num. of mol.: 1 / Mutation: INS(M0), P64A, P65A, INS(K66, D67, E68, L69) Source method: isolated from a genetically manipulated source Details: QAE-COMPONENT / Source: (gene. exp.)  Macrozoarces americanus (ocean pout) / Tissue: BLOOD / Gene: K38 / Variant: HPLC-12 / Plasmid: JM83 / Gene (production host): K38 / Production host: Macrozoarces americanus (ocean pout) / Tissue: BLOOD / Gene: K38 / Variant: HPLC-12 / Plasmid: JM83 / Gene (production host): K38 / Production host:  |
|---|
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR | ||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
|
- Sample preparation
Sample preparation
| Sample conditions | pH: 7.6 / Temperature: 298 K |
|---|---|
| Crystal grow | *PLUS Method: other / Details: NMR |
-NMR measurement
| NMR spectrometer | Type: Varian UNITY / Manufacturer: Varian / Model: UNITY / Field strength: 600 MHz |
|---|
- Processing
Processing
| Software |
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR software |
| ||||||||||||
| Refinement | Method: DG_SUB_EMBED, DGSA, REFINE / Software ordinal: 1 | ||||||||||||
| NMR ensemble | Conformer selection criteria: X-PLOR ENERGIES / Conformers calculated total number: 50 / Conformers submitted total number: 22 |
 Movie
Movie Controller
Controller










 PDBj
PDBj
 X-PLOR
X-PLOR