[English] 日本語
 Yorodumi
Yorodumi- PDB-1ccm: DIRECT NOE REFINEMENT OF CRAMBIN FROM 2D NMR DATA USING A SLOW-CO... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1ccm | ||||||
|---|---|---|---|---|---|---|---|
| Title | DIRECT NOE REFINEMENT OF CRAMBIN FROM 2D NMR DATA USING A SLOW-COOLING ANNEALING PROTOCOL | ||||||
 Components Components | CRAMBIN | ||||||
 Keywords Keywords | PLANT SEED PROTEIN | ||||||
| Function / homology |  Function and homology information Function and homology information | ||||||
| Biological species |  Crambe hispanica subsp. abyssinica (Abyssinian crambe) Crambe hispanica subsp. abyssinica (Abyssinian crambe) | ||||||
| Method | SOLUTION NMR | ||||||
 Authors Authors | Bonvin, A.M.J.J. / Rullmann, J.A.C. / Lamerichs, R.M.J.N. / Boelens, R. / Kaptein, R. | ||||||
 Citation Citation |  Journal: Proteins / Year: 1993 Journal: Proteins / Year: 1993Title: "Ensemble" iterative relaxation matrix approach: a new NMR refinement protocol applied to the solution structure of crambin. Authors: Bonvin, A.M. / Rullmann, J.A. / Lamerichs, R.M. / Boelens, R. / Kaptein, R. #1:  Journal: Proteins / Year: 1993 Journal: Proteins / Year: 1993Title: "Ensemble" Iterative Relaxation Matrix Approach: A New NMR Refinement Protocol Applied to the Solution Structure of Crambin Authors: Bonvin, A.M.J.J. / Rullmann, J.A.C. / Lamerichs, R.M.J.N. / Boelens, R. / Kaptein, R. #2:  Journal: COMPUTATION OF BIOMOLECULAR STRUCTURES; ACHIEVEMENTS PROBLEMS, AND PERSPECTIVES Journal: COMPUTATION OF BIOMOLECULAR STRUCTURES; ACHIEVEMENTS PROBLEMS, AND PERSPECTIVESYear: 1992 Title: Structure Determination by NMR-Application to Crambin Authors: Rullmann, J.A.C. / Bonvin, A.M.J.J. / Boelens, R. / Kaptein, R. #3:  Journal: Thesis / Year: 1989 Journal: Thesis / Year: 1989Title: 2D NMR Studies of Biomolecules: Protein Structure and Protein-DNA Interactions Authors: Lamerichs, R.M.J.M. #4:  Journal: Eur.J.Biochem. / Year: 1988 Journal: Eur.J.Biochem. / Year: 1988Title: Secondary Structure and Hydrogen Bonding of Crambin in Solution Authors: Lamerichs, R.M.J.N. / Berliner, L.J. / Boelens, R. / Demarco, A. / Llinas, M. / Kaptein, R. #5:  Journal: FEBS Lett. / Year: 1987 Journal: FEBS Lett. / Year: 1987Title: 1H NMR Characterization of Two Crambin Species Authors: Vermeulen, J.A.W.H. / Lamerichs, R.M.J.M. / Berliner, L.J. / Demarco, A. / Llinas, M. / Boelens, R. / Alleman, J. / Kaptein, R. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1ccm.cif.gz 1ccm.cif.gz | 55.2 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1ccm.ent.gz pdb1ccm.ent.gz | 36.8 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1ccm.json.gz 1ccm.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/cc/1ccm https://data.pdbj.org/pub/pdb/validation_reports/cc/1ccm ftp://data.pdbj.org/pub/pdb/validation_reports/cc/1ccm ftp://data.pdbj.org/pub/pdb/validation_reports/cc/1ccm | HTTPS FTP |
|---|
-Related structure data
| Related structure data | |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 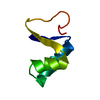
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| Atom site foot note | 1: ARG 17 - LEU 18 OMEGA ANGLE = 147.581 PEPTIDE BOND DEVIATES SIGNIFICANTLY FROM TRANS CONFORMATION IN MODEL 7 | |||||||||
| NMR ensembles |
|
- Components
Components
| #1: Protein/peptide | Mass: 4738.447 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Crambe hispanica subsp. abyssinica (Abyssinian crambe) Crambe hispanica subsp. abyssinica (Abyssinian crambe)Species: Crambe hispanica / Strain: subsp. abyssinica / References: UniProt: P01542 |
|---|---|
| Has protein modification | Y |
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR |
|---|
- Sample preparation
Sample preparation
| Crystal grow | *PLUS Method: other / Details: NMR |
|---|
- Processing
Processing
| NMR software | Name: GROMOS / Developer: VAN GUNSTEREN,BERENDSEN / Classification: refinement |
|---|---|
| NMR ensemble | Conformers submitted total number: 8 |
 Movie
Movie Controller
Controller








 PDBj
PDBj
