[English] 日本語
 Yorodumi
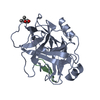
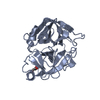
Yorodumi- PDB-1bio: HUMAN COMPLEMENT FACTOR D IN COMPLEX WITH ISATOIC ANHYDRIDE INHIBITOR -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1bio | ||||||
|---|---|---|---|---|---|---|---|
| Title | HUMAN COMPLEMENT FACTOR D IN COMPLEX WITH ISATOIC ANHYDRIDE INHIBITOR | ||||||
 Components Components | COMPLEMENT FACTOR D | ||||||
 Keywords Keywords | SERINE PROTEASE / HYDROLASE / COMPLEMENT / FACTOR D / CATALYTIC TRIAD / SELF-REGULATION | ||||||
| Function / homology |  Function and homology information Function and homology informationcomplement factor D / Alternative complement activation / complement activation / complement activation, alternative pathway / serine-type peptidase activity / platelet alpha granule lumen / protein maturation / response to bacterium / Platelet degranulation / secretory granule lumen ...complement factor D / Alternative complement activation / complement activation / complement activation, alternative pathway / serine-type peptidase activity / platelet alpha granule lumen / protein maturation / response to bacterium / Platelet degranulation / secretory granule lumen / ficolin-1-rich granule lumen / serine-type endopeptidase activity / Neutrophil degranulation / proteolysis / extracellular space / extracellular exosome / extracellular region Similarity search - Function | ||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  MOLECULAR REPLACEMENT / Resolution: 1.5 Å MOLECULAR REPLACEMENT / Resolution: 1.5 Å | ||||||
 Authors Authors | Jing, H. / Babu, Y.S. / Moore, D. / Kilpatrick, J.M. / Liu, X.-Y. / Volanakis, J.E. / Narayana, S.V.L. | ||||||
 Citation Citation |  Journal: J.Mol.Biol. / Year: 1998 Journal: J.Mol.Biol. / Year: 1998Title: Structures of native and complexed complement factor D: implications of the atypical His57 conformation and self-inhibitory loop in the regulation of specific serine protease activity. Authors: Jing, H. / Babu, Y.S. / Moore, D. / Kilpatrick, J.M. / Liu, X.Y. / Volanakis, J.E. / Narayana, S.V. #1:  Journal: Protein Sci. / Year: 1996 Journal: Protein Sci. / Year: 1996Title: Complement Factor D, a Novel Serine Protease Authors: Volanakis, J.E. / Narayana, S.V. #2:  Journal: J.Biol.Chem. / Year: 1995 Journal: J.Biol.Chem. / Year: 1995Title: Crystal Structure of a Complement Factor D Mutant Expressing Enhanced Catalytic Activity Authors: Kim, S. / Narayana, S.V. / Volanakis, J.E. #3:  Journal: J.Biol.Chem. / Year: 1995 Journal: J.Biol.Chem. / Year: 1995Title: Erratum. Crystal Structure of a Complement Factor D Mutant Expressing Enhanced Catalytic Activity Authors: Kim, S. / Narayana, S.V. / Volanakis, J.E. #4:  Journal: J.Mol.Biol. / Year: 1994 Journal: J.Mol.Biol. / Year: 1994Title: Structure of Human Factor D. A Complement System Protein at 2.0 A Resolution Authors: Narayana, S.V. / Carson, M. / El-Kabbani, O. / Kilpatrick, J.M. / Moore, D. / Chen, X. / Bugg, C.E. / Volanakis, J.E. / Delucas, L.J. #5:  Journal: J.Mol.Biol. / Year: 1991 Journal: J.Mol.Biol. / Year: 1991Title: Crystallization and Preliminary X-Ray Investigation of Factor D of Human Complement Authors: Narayana, S.V. / Kilpatrick, J.M. / El-Kabbani, O. / Babu, Y.S. / Bugg, C.E. / Volanakis, J.E. / Delucas, L.J. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1bio.cif.gz 1bio.cif.gz | 63.4 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1bio.ent.gz pdb1bio.ent.gz | 45.2 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1bio.json.gz 1bio.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/bi/1bio https://data.pdbj.org/pub/pdb/validation_reports/bi/1bio ftp://data.pdbj.org/pub/pdb/validation_reports/bi/1bio ftp://data.pdbj.org/pub/pdb/validation_reports/bi/1bio | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  1hfdC  1dsuS S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 24438.807 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Details: ISATOIC ANHYDRIDE ACYLATES ACTIVE SITE SER 195 / Source: (natural)  Homo sapiens (human) / References: UniProt: P00746, complement factor D Homo sapiens (human) / References: UniProt: P00746, complement factor D |
|---|---|
| #2: Chemical | ChemComp-SOA / |
| #3: Chemical | ChemComp-GOL / |
| #4: Water | ChemComp-HOH / |
| Has protein modification | Y |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 2 X-RAY DIFFRACTION / Number of used crystals: 2 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.15 Å3/Da / Density % sol: 44 % | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | pH: 6.4 / Details: pH 6.4 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Crystal grow | *PLUS Method: vapor diffusion, hanging dropDetails: drop consists of equal volume of protein and reservoir solutions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction | Mean temperature: 95 K |
|---|---|
| Diffraction source | Source:  ROTATING ANODE / Type: RIGAKU RUH2R / Wavelength: 1.5418 ROTATING ANODE / Type: RIGAKU RUH2R / Wavelength: 1.5418 |
| Detector | Type: RIGAKU RAXIS IV / Detector: IMAGE PLATE / Date: Jan 1, 1997 / Details: MIRRORS |
| Radiation | Monochromator: GRAPHITE(002) / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1.5418 Å / Relative weight: 1 |
| Reflection | Resolution: 1.5→20 Å / Num. obs: 31552 / % possible obs: 95 % / Observed criterion σ(I): 2 / Redundancy: 4.1 % / Biso Wilson estimate: 17.8 Å2 / Rmerge(I) obs: 0.051 / Net I/σ(I): 29 |
| Reflection shell | Resolution: 1.5→1.55 Å / Redundancy: 4 % / Rmerge(I) obs: 0.253 / Mean I/σ(I) obs: 5.5 / % possible all: 90 |
| Reflection | *PLUS Num. measured all: 133846 |
| Reflection shell | *PLUS % possible obs: 90 % |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB ENTRY 1DSU MOLECULE B Resolution: 1.5→20 Å / Rfactor Rfree error: 0.004 / Data cutoff high absF: 1000000 / Data cutoff low absF: 0.001 / Cross valid method: THROUGHOUT / σ(F): 2 / Details: BULK SOLVENT CORRECTION WAS APPLIED
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 17.3 Å2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine analyze |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.5→20 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 1.5→1.57 Å / Rfactor Rfree error: 0.017 / Total num. of bins used: 8
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Xplor file |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Software | *PLUS Name:  X-PLOR / Version: 3.851 / Classification: refinement X-PLOR / Version: 3.851 / Classification: refinement | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints | *PLUS
|
 Movie
Movie Controller
Controller










 PDBj
PDBj