[English] 日本語
 Yorodumi
Yorodumi- PDB-185d: SEQUENCE SPECIFICITY OF QUINOXALINE ANTIBIOTICS. 1. SOLUTION STRU... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 185d | ||||||
|---|---|---|---|---|---|---|---|
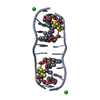
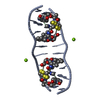
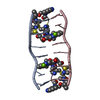
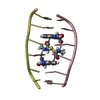
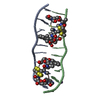
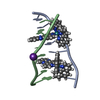
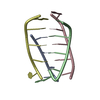
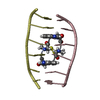
| Title | SEQUENCE SPECIFICITY OF QUINOXALINE ANTIBIOTICS. 1. SOLUTION STRUCTURE OF A 1:1 COMPLEX BETWEEN TRIOSTIN A AND [D(GACGTC)]2 AND COMPARISON WITH THE SOLUTION STRUCTURE OF THE [N-MECYS3, N-MECYS7]TANDEM-[D(GATATC)]2 COMPLEX | ||||||
 Components Components |
| ||||||
 Keywords Keywords | DNA/ANTIBIOTIC / BISINTERCALATOR / DEPSIPEPTIDE / QUINOXALINE / ANTIBIOTIC / ANTITUMOR / DNA-ANTIBIOTIC COMPLEX | ||||||
| Function / homology | TRIOSTIN A / 2-CARBOXYQUINOXALINE / : / DNA Function and homology information Function and homology information | ||||||
| Biological species |  STREPTOMYCINEAE (bacteria) STREPTOMYCINEAE (bacteria) | ||||||
| Method | SOLUTION NMR / DISTANCE GEOMETRY, MOLECULAR DYNAMICS | ||||||
 Authors Authors | Addess, K.J. / Feigon, J. | ||||||
 Citation Citation |  Journal: Biochemistry / Year: 1994 Journal: Biochemistry / Year: 1994Title: Sequence Specificity of Quinoxaline Antibiotics. 1. Solution Structure of a 1:1 Complex between Triostin a and [D(Gacgtc)]2 and Comparison with the Solution Structure of the [N-Mecys3,N-Mecys7] ...Title: Sequence Specificity of Quinoxaline Antibiotics. 1. Solution Structure of a 1:1 Complex between Triostin a and [D(Gacgtc)]2 and Comparison with the Solution Structure of the [N-Mecys3,N-Mecys7]Tandem-[D(Gatatc)]2 Complex. Authors: Addess, K.J. / Feigon, J. #1:  Journal: Biochemistry / Year: 1993 Journal: Biochemistry / Year: 1993Title: Solution Structure of a Complex between [N-Mecys3,N-Mecys7]Tandem and [D(Gatatc)]2 Authors: Addess, K.J. / Sinsheimer, J.S. / Feigon, J. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  185d.cif.gz 185d.cif.gz | 61.7 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb185d.ent.gz pdb185d.ent.gz | 46.4 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  185d.json.gz 185d.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/85/185d https://data.pdbj.org/pub/pdb/validation_reports/85/185d ftp://data.pdbj.org/pub/pdb/validation_reports/85/185d ftp://data.pdbj.org/pub/pdb/validation_reports/85/185d | HTTPS FTP |
|---|
-Related structure data
| Related structure data | |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 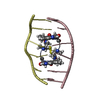
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| Atom site foot note | 1: SER A 2 AND SER B 7 OF EACH MODEL ARE D-SERINE. 2: CYS A 4 AND VAL A 5 OF EACH MODEL ARE METHYLATED. CYS B 9 AND VAL B 10 OF EACH MODEL ARE METHYLATED. | |||||||||
| NMR ensembles |
|
- Components
Components
| #1: Protein/peptide | 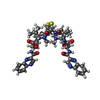  Type: Cyclic depsipeptide / Class: Anticancer / Mass: 794.982 Da / Num. of mol.: 1 / Source method: obtained synthetically Type: Cyclic depsipeptide / Class: Anticancer / Mass: 794.982 Da / Num. of mol.: 1 / Source method: obtained syntheticallyDetails: TRIOSTIN IS A BICYCLIC OCTADEPSIPEPTIDE. BICYCLIZATION IS ACHIEVED BY LINKING THE N- AND THE C- TERMINI, AND A DISULPHIDE BOND BETWEEN RESIDUES 3 AND 7. THE TWO QUINOXALINE CHROMOPHORES ARE ...Details: TRIOSTIN IS A BICYCLIC OCTADEPSIPEPTIDE. BICYCLIZATION IS ACHIEVED BY LINKING THE N- AND THE C- TERMINI, AND A DISULPHIDE BOND BETWEEN RESIDUES 3 AND 7. THE TWO QUINOXALINE CHROMOPHORES ARE LINKED TO THE D-SERINE RESIDUES, RESIDUES 1 AND 5. Source: (synth.)  STREPTOMYCINEAE (bacteria) / STREPTOMYCINEAE (bacteria) /  Keywords: DEOXYRIBONUCLEIC ACID / References: NOR: NOR01129, TRIOSTIN A Keywords: DEOXYRIBONUCLEIC ACID / References: NOR: NOR01129, TRIOSTIN A | ||||||
|---|---|---|---|---|---|---|---|
| #2: DNA chain | Mass: 1809.217 Da / Num. of mol.: 2 / Source method: obtained synthetically #3: Chemical | 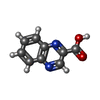  Type: Cyclic depsipeptide / Class: Anticancer / Mass: 174.156 Da / Num. of mol.: 2 / Source method: obtained synthetically / Formula: C9H6N2O2 Type: Cyclic depsipeptide / Class: Anticancer / Mass: 174.156 Da / Num. of mol.: 2 / Source method: obtained synthetically / Formula: C9H6N2O2Details: TRIOSTIN IS A BICYCLIC OCTADEPSIPEPTIDE. BICYCLIZATION IS ACHIEVED BY LINKING THE N- AND THE C- TERMINI, AND A DISULPHIDE BOND BETWEEN RESIDUES 3 AND 7. THE TWO QUINOXALINE CHROMOPHORES ARE ...Details: TRIOSTIN IS A BICYCLIC OCTADEPSIPEPTIDE. BICYCLIZATION IS ACHIEVED BY LINKING THE N- AND THE C- TERMINI, AND A DISULPHIDE BOND BETWEEN RESIDUES 3 AND 7. THE TWO QUINOXALINE CHROMOPHORES ARE LINKED TO THE D-SERINE RESIDUES, RESIDUES 1 AND 5. References: TRIOSTIN A Compound details | TRIOSTIN IS A BICYCLIC OCTADEPSIPEPTIDE, A MEMBER OF THE QUINOXALINE CLASS OF ANTIBIOTICS. HERE, ...TRIOSTIN IS A BICYCLIC OCTADEPSIP | Has protein modification | Y | |
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR |
|---|---|
| NMR details | Text: TRIOSTIN A CONTAINS TWO PLANAR AROMATIC QUINOXALINE RINGS THAT ARE COVALENTLY ATTACHED TO A CYCLIC OCTADEPSIPEPTIDE RING REPRESENTED BY CHAINS A AND B IN THIS ENTRY. THERE IS A DISULFIDE BRIDGE ...Text: TRIOSTIN A CONTAINS TWO PLANAR AROMATIC QUINOXALINE RINGS THAT ARE COVALENTLY ATTACHED TO A CYCLIC OCTADEPSIPEPTIDE RING REPRESENTED BY CHAINS A AND B IN THIS ENTRY. THERE IS A DISULFIDE BRIDGE LINKING CYS A 4 TO CYS B 9. BOTH CYS AND VAL RESIDUES CONTAIN METHYLATED AMIDE NITROGENS; THE TWO D-SER RESIDUES ARE AMIDE BONDED TO THE TWO QUINOXALINE RINGS. |
- Sample preparation
Sample preparation
| Crystal grow | *PLUS Method: other / Details: NMR |
|---|
- Processing
Processing
| Software |
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| NMR software | Name:  X-PLOR / Developer: BRUNGER / Classification: refinement X-PLOR / Developer: BRUNGER / Classification: refinement | ||||||||
| Refinement | Method: DISTANCE GEOMETRY, MOLECULAR DYNAMICS / Software ordinal: 1 Details: NUMBER OF ATOMS PRESENT IN ENTRY. NUMBER OF PROTEIN ATOMS 328 NUMBER OF NUCLEIC ACID ATOMS 48 NUMBER OF SOLVENT ATOMS 240 NUMBER OF HETEROGEN ATOMS 40 AVERAGE PAIRWISE RMSD BOND DISTANCES ...Details: NUMBER OF ATOMS PRESENT IN ENTRY. NUMBER OF PROTEIN ATOMS 328 NUMBER OF NUCLEIC ACID ATOMS 48 NUMBER OF SOLVENT ATOMS 240 NUMBER OF HETEROGEN ATOMS 40 AVERAGE PAIRWISE RMSD BOND DISTANCES FOR HEAVY ATOM POSITIONS FOR 5 STRUCTURES: 1.37 ANGSTROMS. | ||||||||
| NMR ensemble | Conformers submitted total number: 5 |
 Movie
Movie Controller
Controller










 PDBj
PDBj