+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1pfe | ||||||
|---|---|---|---|---|---|---|---|
| Title | Echinomycin-(gcgtacgc)2 complex | ||||||
 Components Components |
| ||||||
 Keywords Keywords | DNA/ANTIBIOTIC / BISINTERCALATOR / HOOGSTEEN BASEPAIR / DEPSIPEPTIDE / QUINOXALINE / THIOACETAL / ANTIBIOTIC / ANTITUMOR / DNA-ANTIBIOTIC COMPLEX | ||||||
| Function / homology | Echinomycin / 2-CARBOXYQUINOXALINE / : / DNA Function and homology information Function and homology information | ||||||
| Biological species |  STREPTOMYCES ECHINATUS (bacteria) STREPTOMYCES ECHINATUS (bacteria) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.1 Å MOLECULAR REPLACEMENT / Resolution: 1.1 Å | ||||||
 Authors Authors | Cuesta-Seijo, J.A. | ||||||
 Citation Citation |  Journal: Acta Crystallogr.,Sect.D / Year: 2005 Journal: Acta Crystallogr.,Sect.D / Year: 2005Title: Structures of Complexes between Echinomycin and Duplex DNA. Authors: Cuesta-Seijo, J.A. / Sheldrick, G.M. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1pfe.cif.gz 1pfe.cif.gz | 30.3 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1pfe.ent.gz pdb1pfe.ent.gz | 20.2 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1pfe.json.gz 1pfe.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/pf/1pfe https://data.pdbj.org/pub/pdb/validation_reports/pf/1pfe ftp://data.pdbj.org/pub/pdb/validation_reports/pf/1pfe ftp://data.pdbj.org/pub/pdb/validation_reports/pf/1pfe | HTTPS FTP |
|---|
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| |||||||||||||||
| Unit cell |
| |||||||||||||||
| Components on special symmetry positions |
| |||||||||||||||
| Details | The second part of the biological assembly is generated by: -x, -x+y, -z |
- Components
Components
| #1: DNA chain | Mass: 2427.605 Da / Num. of mol.: 1 / Source method: obtained synthetically | ||||
|---|---|---|---|---|---|
| #2: Protein/peptide |   Type: Cyclic depsipeptide / Class: Antibiotic / Mass: 809.008 Da / Num. of mol.: 1 / Source method: isolated from a natural source Type: Cyclic depsipeptide / Class: Antibiotic / Mass: 809.008 Da / Num. of mol.: 1 / Source method: isolated from a natural sourceDetails: ECHINOMYCIN IS A BICYCLIC OCTADEPSIPEPTIDE. BICYCLIZATION IS ACHIEVED BY LINKING THE N- AND THE C- TERMINI, AND A THIOACETAL BOND BETWEEN RESIDUES 3 AND 7. THE TWO QUINOXALINE CHROMOPHORES ...Details: ECHINOMYCIN IS A BICYCLIC OCTADEPSIPEPTIDE. BICYCLIZATION IS ACHIEVED BY LINKING THE N- AND THE C- TERMINI, AND A THIOACETAL BOND BETWEEN RESIDUES 3 AND 7. THE TWO QUINOXALINE CHROMOPHORES ARE LINKED TO THE D-SERINE RESIDUES, RESIDUES 1 AND 5. Source: (natural)  STREPTOMYCES ECHINATUS (bacteria) / References: NOR: NOR01126, Echinomycin STREPTOMYCES ECHINATUS (bacteria) / References: NOR: NOR01126, Echinomycin | ||||
| #3: Chemical | ChemComp-CL / | ||||
| #4: Chemical | 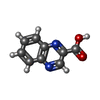  Type: Cyclic depsipeptide / Class: Antibiotic / Mass: 174.156 Da / Num. of mol.: 2 / Source method: obtained synthetically / Formula: C9H6N2O2 Type: Cyclic depsipeptide / Class: Antibiotic / Mass: 174.156 Da / Num. of mol.: 2 / Source method: obtained synthetically / Formula: C9H6N2O2Details: ECHINOMYCIN IS A BICYCLIC OCTADEPSIPEPTIDE. BICYCLIZATION IS ACHIEVED BY LINKING THE N- AND THE C- TERMINI, AND A THIOACETAL BOND BETWEEN RESIDUES 3 AND 7. THE TWO QUINOXALINE CHROMOPHORES ...Details: ECHINOMYCIN IS A BICYCLIC OCTADEPSIPEPTIDE. BICYCLIZATION IS ACHIEVED BY LINKING THE N- AND THE C- TERMINI, AND A THIOACETAL BOND BETWEEN RESIDUES 3 AND 7. THE TWO QUINOXALINE CHROMOPHORES ARE LINKED TO THE D-SERINE RESIDUES, RESIDUES 1 AND 5. References: Echinomycin #5: Water | ChemComp-HOH / | Compound details | THE ECHINOMYCIN IS A BICYCLIC OCTADEPSIPEPTIDE, A MEMBER OF THE QUINOXALINE CLASS OF ANTIBIOTICS. ...THE ECHINOMYCI | |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 3.51 Å3/Da / Density % sol: 64.98 % | ||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | pH: 4.5 Details: LI2SO4, MGCL2, METHANOL, NAAC, SPERMINE, PH 4.5, VAPOR DIFFUSION, HANGING DROP, TEMPERATURE 293K | ||||||||||||||||||||||||||||||||
| Components of the solutions |
|
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  EMBL/DESY, HAMBURG EMBL/DESY, HAMBURG  / Beamline: X11 / Wavelength: 0.8162 / Beamline: X11 / Wavelength: 0.8162 |
| Detector | Type: MARRESEARCH / Detector: CCD / Date: Nov 19, 2002 |
| Radiation | Monochromator: TRIANGULAR MONOCHROMATOR / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.8162 Å / Relative weight: 1 |
| Reflection | Resolution: 1.1→21 Å / Num. obs: 15164 / % possible obs: 97.2 % / Observed criterion σ(I): 0 / Redundancy: 14.91 % / Rmerge(I) obs: 0.049 / Net I/σ(I): 30.01 |
| Reflection shell | Resolution: 1.1→1.2 Å / Redundancy: 13.05 % / Rmerge(I) obs: 0.242 / Mean I/σ(I) obs: 11.48 / % possible all: 95.7 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: TROSTIN A - (GCGTACGC)2 COMPLEX Resolution: 1.1→21 Å / Num. parameters: 2979 / Num. restraintsaints: 2074 / Cross valid method: FREE R / σ(F): 0 StereochEM target val spec case: TARGETS FOR ECHINOMYCIN BUILT FROM THE CSD Stereochemistry target values: STANDARD SHELX FOR THE DNA, ENGH & HUBER FOR THE STANDARD PROTEIN PARTS OF ECHINOMYCIN, BUILT FROM THE CSD FOR THE REST Details: THE SKELETON OF ECHINOMYCIN IS NEARLY SYMMETRIC (NOT MODELLED TO DO SO), THE BRIDGE IS NOT. BOTH POSSIBLE ORIENTATIONS OF BINDING WERE OBSERVED, BUT SKELETON POSITIONS NEARLY OVERLAP, SO ...Details: THE SKELETON OF ECHINOMYCIN IS NEARLY SYMMETRIC (NOT MODELLED TO DO SO), THE BRIDGE IS NOT. BOTH POSSIBLE ORIENTATIONS OF BINDING WERE OBSERVED, BUT SKELETON POSITIONS NEARLY OVERLAP, SO ONLY THE BRIDGE WAS MODELLED AS DISORDERED/MICROHETEROGENEITY.
| |||||||||||||||||||||||||||||||||
| Refine analyze | Num. disordered residues: 7 / Occupancy sum hydrogen: 149 / Occupancy sum non hydrogen: 303 | |||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.1→21 Å
| |||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 1.1→1.2 Å / % reflection obs: 95.7 % |
 Movie
Movie Controller
Controller









 PDBj
PDBj


