[English] 日本語
 Yorodumi
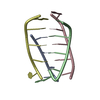
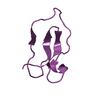
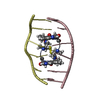
Yorodumi- PDB-1i25: Three dimensional solution structure of huwentoxin-II by 2D 1H-NMR -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1i25 | ||||||
|---|---|---|---|---|---|---|---|
| Title | Three dimensional solution structure of huwentoxin-II by 2D 1H-NMR | ||||||
 Components Components | HUWENTOXIN-II | ||||||
 Keywords Keywords | TOXIN / neurotoxin / insecticidal toxin / disulfide bonds | ||||||
| Function / homology | Huwentoxin-II family / Huwentoxin-II family / Huwentoxin-2 family signature. / host cell postsynaptic membrane / toxin activity / extracellular region / U1-theraphotoxin-Hs1a Function and homology information Function and homology information | ||||||
| Biological species |  Ornithoctonus huwena (Chinese earth tiger) Ornithoctonus huwena (Chinese earth tiger) | ||||||
| Method | SOLUTION NMR / simulated annealing | ||||||
 Authors Authors | Shu, Q. / Lu, S.Y. / Gu, X.C. / Liang, S.P. | ||||||
 Citation Citation |  Journal: Protein Sci. / Year: 2002 Journal: Protein Sci. / Year: 2002Title: The structure of spider toxin huwentoxin-II with unique disulfide linkage: evidence for structural evolution. Authors: Shu, Q. / Lu, S.Y. / Gu, X.C. / Liang, S.P. #1:  Journal: ACTA BIOCHIM.BIOPHYS.SINICA / Year: 2001 Journal: ACTA BIOCHIM.BIOPHYS.SINICA / Year: 2001Title: Sequence-specific Assignment of 1H-NMR Resonance and Determination of the Secondary Structure of HWTX-II Authors: Shu, Q. / Lu, S.Y. / Gu, X.C. / Liang, S.P. #2:  Journal: To be Published Journal: To be PublishedTitle: Assignment of Disulfide Bonds of Huwentoxin-II by Edman Degradation Sequencing and Stepwise Thiol Modification Authors: Shu, Q. / Huang, R.H. / Liang, S.P. #3:  Journal: J.PEPT.RES. / Year: 1999 Journal: J.PEPT.RES. / Year: 1999Title: Purification and Characterization of Huwentoxin-II, a Neurotoxic Peptide from the Venom of the Chinese Bird Spider Selenocosmia huwena Authors: Shu, Q. / Liang, S.P. | ||||||
| History |
| ||||||
| Remark 750 | TURN DETERMINATION METHOD: AUTHOR-DETERMINED |
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1i25.cif.gz 1i25.cif.gz | 120.9 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1i25.ent.gz pdb1i25.ent.gz | 98.4 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1i25.json.gz 1i25.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/i2/1i25 https://data.pdbj.org/pub/pdb/validation_reports/i2/1i25 ftp://data.pdbj.org/pub/pdb/validation_reports/i2/1i25 ftp://data.pdbj.org/pub/pdb/validation_reports/i2/1i25 | HTTPS FTP |
|---|
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: Protein/peptide | Mass: 4300.246 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Details: VENOM Source: (natural)  Ornithoctonus huwena (Chinese earth tiger) Ornithoctonus huwena (Chinese earth tiger)Organ: VENOM GLAND / References: UniProt: P82959 |
|---|---|
| Has protein modification | Y |
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
| ||||||||||||||||||||||||||||||||||||
| NMR details | Text: This structure was determined using standard 2D homonuclear techniques. |
- Sample preparation
Sample preparation
| Details |
| |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample conditions |
| |||||||||||||||
| Crystal grow | *PLUS Method: other / Details: NMR |
-NMR measurement
| NMR spectrometer | Type: Bruker DMX / Manufacturer: Bruker / Model: DMX / Field strength: 500 MHz |
|---|
- Processing
Processing
| NMR software |
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method: simulated annealing / Software ordinal: 1 Details: The structures are based on 592 NOE-derived distance constraints, 16 dihedral angel restraints and 9 fake distance restraints from disulfide bonds. | ||||||||||||
| NMR representative | Selection criteria: closest to the average | ||||||||||||
| NMR ensemble | Conformer selection criteria: structures with the lowest energy Conformers calculated total number: 50 / Conformers submitted total number: 10 |
 Movie
Movie Controller
Controller









 PDBj
PDBj X-PLOR
X-PLOR