[English] 日本語
 Yorodumi
Yorodumi- EMDB-9731: Structure of a substrate engaged SecA-SecY protein translocation ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-9731 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of a substrate engaged SecA-SecY protein translocation machine | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | SecA / SecY / Translocation / Cryo-EM / PROTEIN TRANSPORT | |||||||||
| Function / homology |  Function and homology information Function and homology informationouter membrane protein complex / cell envelope Sec protein transport complex / protein-exporting ATPase activity / protein-secreting ATPase / monoatomic ion transmembrane transporter activity / protein transport by the Sec complex / intracellular protein transmembrane transport / detection of virus / outer membrane / protein import ...outer membrane protein complex / cell envelope Sec protein transport complex / protein-exporting ATPase activity / protein-secreting ATPase / monoatomic ion transmembrane transporter activity / protein transport by the Sec complex / intracellular protein transmembrane transport / detection of virus / outer membrane / protein import / porin activity / pore complex / protein secretion / protein transmembrane transporter activity / protein targeting / monoatomic ion transport / bioluminescence / generation of precursor metabolites and energy / cell outer membrane / outer membrane-bounded periplasmic space / membrane raft / DNA damage response / symbiont entry into host cell / ATP binding / metal ion binding / identical protein binding / membrane / plasma membrane / cytoplasm Similarity search - Function | |||||||||
| Biological species |    Geobacillus thermodenitrificans (strain NG80-2) (bacteria) / Geobacillus thermodenitrificans (strain NG80-2) (bacteria) /    | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.45 Å | |||||||||
 Authors Authors | Ma CY / Wu XF | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2019 Journal: Nat Commun / Year: 2019Title: Structure of the substrate-engaged SecA-SecY protein translocation machine. Authors: Chengying Ma / Xiaofei Wu / Dongjie Sun / Eunyong Park / Marco A Catipovic / Tom A Rapoport / Ning Gao / Long Li /   Abstract: The Sec61/SecY channel allows the translocation of many proteins across the eukaryotic endoplasmic reticulum membrane or the prokaryotic plasma membrane. In bacteria, most secretory proteins are ...The Sec61/SecY channel allows the translocation of many proteins across the eukaryotic endoplasmic reticulum membrane or the prokaryotic plasma membrane. In bacteria, most secretory proteins are transported post-translationally through the SecY channel by the SecA ATPase. How a polypeptide is moved through the SecA-SecY complex is poorly understood, as structural information is lacking. Here, we report an electron cryo-microscopy (cryo-EM) structure of a translocating SecA-SecY complex in a lipid environment. The translocating polypeptide chain can be traced through both SecA and SecY. In the captured transition state of ATP hydrolysis, SecA's two-helix finger is close to the polypeptide, while SecA's clamp interacts with the polypeptide in a sequence-independent manner by inducing a short β-strand. Taking into account previous biochemical and biophysical data, our structure is consistent with a model in which the two-helix finger and clamp cooperate during the ATPase cycle to move a polypeptide through the channel. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_9731.map.gz emd_9731.map.gz | 3.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-9731-v30.xml emd-9731-v30.xml emd-9731.xml emd-9731.xml | 22.9 KB 22.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_9731.png emd_9731.png | 55.7 KB | ||
| Filedesc metadata |  emd-9731.cif.gz emd-9731.cif.gz | 8.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-9731 http://ftp.pdbj.org/pub/emdb/structures/EMD-9731 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9731 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9731 | HTTPS FTP |
-Related structure data
| Related structure data |  6itcMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_9731.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_9731.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.05 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
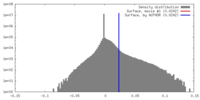
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
+Entire : SecA-SecY complex
+Supramolecule #1: SecA-SecY complex
+Macromolecule #1: Protein translocase subunit SecA
+Macromolecule #2: Protein translocase subunit SecY
+Macromolecule #3: Protein translocase subunit SecE
+Macromolecule #4: Nanobody
+Macromolecule #5: Translocating peptide
+Macromolecule #6: Green fluorescent protein
+Macromolecule #7: Nanobody
+Macromolecule #8: MAGNESIUM ION
+Macromolecule #9: BERYLLIUM TRIFLUORIDE ION
+Macromolecule #10: ADENOSINE-5'-DIPHOSPHATE
+Macromolecule #11: (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-...
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Average electron dose: 5.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: FLEXIBLE FIT |
|---|---|
| Output model |  PDB-6itc: |
 Movie
Movie Controller
Controller
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)