+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-9244 | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
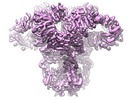
| Title | Structure of full-length IP3R1 channel in Apo-state (composite) | |||||||||||||||||||||
 Map data Map data | cryoEM structure of IP3R1 in an apo state | |||||||||||||||||||||
 Sample Sample |
| |||||||||||||||||||||
 Keywords Keywords | inositol 1 / 4 / 5-trisphosphate receptor / calcium release channel / neuronal type 1 / MEMBRANE PROTEIN | |||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationEffects of PIP2 hydrolysis / Antigen activates B Cell Receptor (BCR) leading to generation of second messengers / inositol 1,4,5-trisphosphate receptor activity involved in regulation of postsynaptic cytosolic calcium levels / release of sequestered calcium ion into cytosol by endoplasmic reticulum / smooth endoplasmic reticulum membrane / cGMP effects / Elevation of cytosolic Ca2+ levels / platelet dense tubular network / calcineurin complex / platelet dense granule membrane ...Effects of PIP2 hydrolysis / Antigen activates B Cell Receptor (BCR) leading to generation of second messengers / inositol 1,4,5-trisphosphate receptor activity involved in regulation of postsynaptic cytosolic calcium levels / release of sequestered calcium ion into cytosol by endoplasmic reticulum / smooth endoplasmic reticulum membrane / cGMP effects / Elevation of cytosolic Ca2+ levels / platelet dense tubular network / calcineurin complex / platelet dense granule membrane / epithelial fluid transport / inositol 1,4,5-trisphosphate-gated calcium channel activity / phospholipase C-activating G protein-coupled acetylcholine receptor signaling pathway / calcium import into the mitochondrion / voluntary musculoskeletal movement / inositol 1,4,5 trisphosphate binding / positive regulation of hepatocyte proliferation / negative regulation of calcium-mediated signaling / positive regulation of calcium ion transport / endoplasmic reticulum calcium ion homeostasis / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / nuclear inner membrane / Ion homeostasis / transport vesicle membrane / dendrite development / intracellularly gated calcium channel activity / ligand-gated ion channel signaling pathway / intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress / regulation of cytosolic calcium ion concentration / single fertilization / calcium channel inhibitor activity / release of sequestered calcium ion into cytosol / phosphatidylinositol binding / liver regeneration / secretory granule membrane / cellular response to cAMP / synaptic membrane / sarcoplasmic reticulum / post-embryonic development / positive regulation of neuron projection development / positive regulation of insulin secretion / GABA-ergic synapse / Schaffer collateral - CA1 synapse / cell morphogenesis / calcium ion transport / nuclear envelope / presynapse / positive regulation of cytosolic calcium ion concentration / protein phosphatase binding / cellular response to hypoxia / phospholipase C-activating G protein-coupled receptor signaling pathway / protein homotetramerization / transmembrane transporter binding / response to hypoxia / postsynapse / postsynaptic density / positive regulation of apoptotic process / protein domain specific binding / neuronal cell body / calcium ion binding / synapse / dendrite / endoplasmic reticulum membrane / negative regulation of apoptotic process / protein-containing complex binding / nucleolus / perinuclear region of cytoplasm / endoplasmic reticulum / protein homodimerization activity / protein-containing complex / ATP binding / identical protein binding / membrane / plasma membrane / cytoplasm Similarity search - Function | |||||||||||||||||||||
| Biological species |  | |||||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.9 Å | |||||||||||||||||||||
 Authors Authors | Serysheva II / Fan G / Baker MR / Wang Z / Seryshev A / Ludtke SJ / Baker ML | |||||||||||||||||||||
| Funding support |  United States, 6 items United States, 6 items
| |||||||||||||||||||||
 Citation Citation |  Journal: Cell Res / Year: 2018 Journal: Cell Res / Year: 2018Title: Cryo-EM reveals ligand induced allostery underlying InsPR channel gating. Authors: Guizhen Fan / Mariah R Baker / Zhao Wang / Alexander B Seryshev / Steven J Ludtke / Matthew L Baker / Irina I Serysheva /  Abstract: Inositol-1,4,5-trisphosphate receptors (InsPRs) are cation channels that mobilize Ca from intracellular stores in response to a wide range of cellular stimuli. The paradigm of InsPR activation is the ...Inositol-1,4,5-trisphosphate receptors (InsPRs) are cation channels that mobilize Ca from intracellular stores in response to a wide range of cellular stimuli. The paradigm of InsPR activation is the coupled interplay between binding of InsP and Ca that switches the ion conduction pathway between closed and open states to enable the passage of Ca through the channel. However, the molecular mechanism of how the receptor senses and decodes ligand-binding signals into gating motion remains unknown. Here, we present the electron cryo-microscopy structure of InsPR1 from rat cerebellum determined to 4.1 Å resolution in the presence of activating concentrations of Ca and adenophostin A (AdA), a structural mimetic of InsP and the most potent known agonist of the channel. Comparison with the 3.9 Å-resolution structure of InsPR1 in the Apo-state, also reported herein, reveals the binding arrangement of AdA in the tetrameric channel assembly and striking ligand-induced conformational rearrangements within cytoplasmic domains coupled to the dilation of a hydrophobic constriction at the gate. Together, our results provide critical insights into the mechanistic principles by which ligand-binding allosterically gates InsPR channel. | |||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_9244.map.gz emd_9244.map.gz | 28 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-9244-v30.xml emd-9244-v30.xml emd-9244.xml emd-9244.xml | 18 KB 18 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_9244.png emd_9244.png | 224.7 KB | ||
| Filedesc metadata |  emd-9244.cif.gz emd-9244.cif.gz | 8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-9244 http://ftp.pdbj.org/pub/emdb/structures/EMD-9244 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9244 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9244 | HTTPS FTP |
-Related structure data
| Related structure data |  6mu2MC  9243C  9245C  9246C  9247C  9248C  6mu1C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_9244.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_9244.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | cryoEM structure of IP3R1 in an apo state | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.26 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Inositol 1,4,5-trisphosphate receptor
| Entire | Name: Inositol 1,4,5-trisphosphate receptor |
|---|---|
| Components |
|
-Supramolecule #1: Inositol 1,4,5-trisphosphate receptor
| Supramolecule | Name: Inositol 1,4,5-trisphosphate receptor / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all / Details: tetrameric assembly |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 1.3 MDa |
-Macromolecule #1: Inositol 1,4,5-trisphosphate receptor type 1
| Macromolecule | Name: Inositol 1,4,5-trisphosphate receptor type 1 / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 313.657406 KDa |
| Sequence | String: MSDKMSSFLH IGDICSLYAE GSTNGFISTL GLVDDRCVVQ PEAGDLNNPP KKFRDCLFKL CPMNRYSAQK QFWKAAKPGA NSTTDAVLL NKLHHAADLE KKQNETENRK LLGTVIQYGN VIQLLHLKSN KYLTVNKRLP ALLEKNAMRV TLDEAGNEGS W FYIQPFYK ...String: MSDKMSSFLH IGDICSLYAE GSTNGFISTL GLVDDRCVVQ PEAGDLNNPP KKFRDCLFKL CPMNRYSAQK QFWKAAKPGA NSTTDAVLL NKLHHAADLE KKQNETENRK LLGTVIQYGN VIQLLHLKSN KYLTVNKRLP ALLEKNAMRV TLDEAGNEGS W FYIQPFYK LRSIGDSVVI GDKVVLNPVN AGQPLHASSH QLVDNPGCNE VNSVNCNTSW KIVLFMKWSD NKDDILKGGD VV RLFHAEQ EKFLTCDEHR KKQHVFLRTT GRQSATSATS SKALWEVEVV QHDPCRGGAG YWNSLFRFKH LATGHYLAAE VDP DFEEEC LEFQPSVDPD QDASRSRLRN AQEKMVYSLV SVPEGNDISS IFELDPTTLR GGDSLVPRNS YVRLRHLCTN TWVH STNIP IDKEEEKPVM LKIGTSPLKE DKEAFAIVPV SPAEVRDLDF ANDASKVLGS IAGKLEKGTI TQNERRSVTK LLEDL VYFV TGGTNSGQDV LEVVFSKPNR ERQKLMREQN ILKQIFKLLQ APFTDCGDGP MLRLEELGDQ RHAPFRHICR LCYRVL RHS QQDYRKNQEY IAKQFGFMQK QIGYDVLAED TITALLHNNR KLLEKHITAA EIDTFVSLVR KNREPRFLDY LSDLCVS MN KSIPVTQELI CKAVLNPTNA DILIETKLVL SRFEFEGVST GENALEAGED EEEVWLFWRD SNKEIRSKSV RELAQDAK E GQKEDRDVLS YYRYQLNLFA RMCLDRQYLA INEISGQLDV DLILRCMSDE NLPYDLRASF CRLMLHMHVD RDPQEQVTP VKYARLWSEI PSEIAIDDYD SSGASKDEIK ERFAQTMEFV EEYLRDVVCQ RFPFSDKEKN KLTFEVVNLA RNLIYFGFYN FSDLLRLTK ILLAILDCVH VTTIFPISKM TKGEENKGSN VMRSIHGVGE LMTQVVLRGG GFLPMTPMAA APEGNVKQAE P EKEDIMVM DTKLKIIEIL QFILNVRLDY RISCLLCIFK REFDESNSQS SETSSGNSSQ EGPSNVPGAL DFEHIEEQAE GI FGGSEEN TPLDLDDHGG RTFLRVLLHL TMHDYPPLVS GALQLLFRHF SQRQEVLQAF KQVQLLVTSQ DVDNYKQIKQ DLD QLRSIV EKSELWVYKG QGPDEPMDGA SGENEHKKTE EGTSKPLKHE STSSYNYRVV KEILIRLSKL CVQESASVRK SRKQ QQRLL RNMGAHAVVL ELLQIPYEKA EDTKMQEIMR LAHEFLQNFC AGNQQNQALL HKHINLFLNP GILEAVTMQH IFMNN FQLC SEINERVVQH FVHCIETHGR NVQYIKFLQT IVKAEGKFIK KCQDMVMAEL VNSGEDVLVF YNDRASFQTL IQMMRS ERD RMDENSPLFM YHIHLVELLA VCTEGKNVYT EIKCNSLLPL DDIVRVVTHE DCIPEVKIAY INFLNHCYVD TEVEMKE IY TSNHMWKLFE NFLVDICRAC NNTSDRKHAD SVLEKYVTEI VMSIVTTFFS SPFSDQSTTL QTRQPVFVQL LQGVFRVY H CNWLMPSQKA SVESCIRVLS DVAKSRAIAI PVDLDSQVNN LFLKSHNIVQ KTAMNWRLSA RNAARRDSVL AASRDYRNI IERLQDIVSA LEDRLRPLVQ AELSVLVDVL HRPELLFPEN TDARRKCESG GFICKLIKHT KQLLEENEEK LCIKVLQTLR EMMTKDRGY GEKQISIDEL ENAELPQPPE AENSTEQELE PSPPLRQLED HKRGEALRQI LVNRYYGNIR PSGRRESLTS F GNGPLSPG GPSKPGGGGG GPGSGSTSRG EMSLAEVQCH LDKEGASNLV IDLIMNASSD RVFHESILLA IALLEGGNTT IQ HSFFCRL TEDKKSEKFF KVFYDRMKVA QQEIKATVTV NTSDLGNKKK DDEVDRDAPS RKKAKEPTTQ ITEEVRDQLL EAS AATRKA FTTFRREADP DDHYQSGEGT QATTDKAKDD LEMSAVITIM QPILRFLQLL CENHNRDLQN FLRCQNNKTN YNLV CETLQ FLDCICGSTT GGLGLLGLYI NEKNVALINQ TLESLTEYCQ GPCHENQNCI ATHESNGIDI ITALILNDIN PLGKK RMDL VLELKNNASK LLLAIMESRH DSENAERILY NMRPKELVEV IKKAYMQGEV EFEDGENGED GAASPRNVGH NIYILA HQL ARHNKELQTM LKPGGQVDGD EALEFYAKHT AQIEIVRLDR TMEQIVFPVP SICEFLTKES KLRIYYTTER DEQGSKI ND FFLRSEDLFN EMNWQKKLRA QPVLYWCARN MSFWSSISFN LAVLMNLLVA FFYPFKGVRG GTLEPHWSGL LWTAMLIS L AIVIALPKPH GIRALIASTI LRLIFSVGLQ PTLFLLGAFN VCNKIIFLMS FVGNCGTFTR GYRAMVLDVE FLYHLLYLL ICAMGLFVHE FFYSLLLFDL VYREETLLNV IKSVTRNGRP IILTAALALI LVYLFSIVGY LFFKDDFILE VDRLPNETAG PETGESLAN DFLYSDVCRV ETGENCTSPA PKEELLPVEE TEQDKEHTCE TLLMCIVTVL SHGLRSGGGV GDVLRKPSKE E PLFAARVI YDLLFFFMVI IIVLNLIFGV IIDTFADLRS EKQKKEEILK TTCFICGLER DKFDNKTVTF EEHIKEEHNM WH YLCFIVL VKVKDSTEYT GPESYVAEMI RERNLDWFPR MRAMSLVSSD SEGEQNELRN LQEKLESTMK LVTNLSGQLS ELK DQMTEQ RKQKQRIGLL GHPPHMNVNP QQPA UniProtKB: Inositol 1,4,5-trisphosphate-gated calcium channel ITPR1 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.1 mg/mL |
|---|---|
| Buffer | pH: 7.4 Details: 50 mM Tris-HCl buffer (pH 7.4), 150 mM NaCl, 1 mM DTT, 0.4% CHAPS, 2 mM EGTA, 1 mM EDTA, protease inhibitors |
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 10 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 293 K / Instrument: FEI VITROBOT MARK IV |
| Details | Monodisperse sample |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F30 |
|---|---|
| Temperature | Min: 93.0 K / Max: 93.0 K |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Frames/image: 2-17 / Number real images: 9823 / Average exposure time: 0.2 sec. / Average electron dose: 1.3 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Sample stage | Specimen holder model: GATAN 910 MULTI-SPECIMEN SINGLE TILT CRYO TRANSFER HOLDER Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Tecnai F30 / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-6mu2: |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)