[English] 日本語
 Yorodumi
Yorodumi- EMDB-20849: Cryo-EM structure of type 3 IP3 receptor revealing presence of a ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-20849 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of type 3 IP3 receptor revealing presence of a self-binding peptide | |||||||||
 Map data Map data | final map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | inositol trisphosphate receptor / InsP3R / IP3R / cryoelectron microscopy / ion channel / calcium channel / isothermal titration calorimetry / self binding peptide / TRANSPORT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationDAG and IP3 signaling / inositol 1,3,4,5 tetrakisphosphate binding / sensory perception of bitter taste / inositol 1,4,5-trisphosphate-gated calcium channel activity / platelet dense tubular network membrane / sensory perception of umami taste / Effects of PIP2 hydrolysis / sensory perception of sweet taste / Elevation of cytosolic Ca2+ levels / PLC beta mediated events ...DAG and IP3 signaling / inositol 1,3,4,5 tetrakisphosphate binding / sensory perception of bitter taste / inositol 1,4,5-trisphosphate-gated calcium channel activity / platelet dense tubular network membrane / sensory perception of umami taste / Effects of PIP2 hydrolysis / sensory perception of sweet taste / Elevation of cytosolic Ca2+ levels / PLC beta mediated events / inositol 1,4,5 trisphosphate binding / inositol hexakisphosphate binding / CLEC7A (Dectin-1) induces NFAT activation / transport vesicle membrane / cytoplasmic side of endoplasmic reticulum membrane / intracellularly gated calcium channel activity / nuclear outer membrane / brush border / Role of phospholipids in phagocytosis / calcium ion homeostasis / Ion homeostasis / release of sequestered calcium ion into cytosol / FCERI mediated Ca+2 mobilization / phosphatidylinositol binding / FCGR3A-mediated IL10 synthesis / Antigen activates B Cell Receptor (BCR) leading to generation of second messengers / secretory granule membrane / VEGFR2 mediated cell proliferation / sarcoplasmic reticulum / Regulation of insulin secretion / response to calcium ion / platelet activation / memory / apical part of cell / Sensory perception of sweet, bitter, and umami (glutamate) taste / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / long-term synaptic potentiation / sensory perception of taste / positive regulation of cytosolic calcium ion concentration / Ca2+ pathway / protein homotetramerization / receptor complex / G protein-coupled receptor signaling pathway / neuronal cell body / calcium ion binding / endoplasmic reticulum membrane / nucleolus / endoplasmic reticulum / zinc ion binding / nucleoplasm / ATP binding / membrane / plasma membrane / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.77 Å | |||||||||
 Authors Authors | Azumaya CM / Linton EA | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: J Biol Chem / Year: 2020 Journal: J Biol Chem / Year: 2020Title: Cryo-EM structure of human type-3 inositol triphosphate receptor reveals the presence of a self-binding peptide that acts as an antagonist. Authors: Caleigh M Azumaya / Emily A Linton / Caitlin J Risener / Terunaga Nakagawa / Erkan Karakas /  Abstract: Calcium-mediated signaling through inositol 1,4,5-triphosphate receptors (IPRs) is essential for the regulation of numerous physiological processes, including fertilization, muscle contraction, ...Calcium-mediated signaling through inositol 1,4,5-triphosphate receptors (IPRs) is essential for the regulation of numerous physiological processes, including fertilization, muscle contraction, apoptosis, secretion, and synaptic plasticity. Deregulation of IPRs leads to pathological calcium signaling and is implicated in many common diseases, including cancer and neurodegenerative, autoimmune, and metabolic diseases. Revealing the mechanism of activation and inhibition of this ion channel will be critical to an improved understanding of the biological processes that are controlled by IPRs. Here, we report structural findings of the human type-3 IPR (IPR-3) obtained by cryo-EM (at an overall resolution of 3.8 Å), revealing an unanticipated regulatory mechanism where a loop distantly located in the primary sequence occupies the IP-binding site and competitively inhibits IP binding. We propose that this inhibitory mechanism must differ qualitatively among IPR subtypes because of their diverse loop sequences, potentially serving as a key molecular determinant of subtype-specific calcium signaling in IPRs. In summary, our structural characterization of human IPR-3 provides critical insights into the mechanistic function of IPRs and into subtype-specific regulation of these important calcium-regulatory channels. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_20849.map.gz emd_20849.map.gz | 139.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-20849-v30.xml emd-20849-v30.xml emd-20849.xml emd-20849.xml | 21.5 KB 21.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_20849.png emd_20849.png | 85.7 KB | ||
| Masks |  emd_20849_msk_1.map emd_20849_msk_1.map | 149.9 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-20849.cif.gz emd-20849.cif.gz | 7.6 KB | ||
| Others |  emd_20849_half_map_1.map.gz emd_20849_half_map_1.map.gz emd_20849_half_map_2.map.gz emd_20849_half_map_2.map.gz | 49 MB 49 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-20849 http://ftp.pdbj.org/pub/emdb/structures/EMD-20849 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20849 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20849 | HTTPS FTP |
-Related structure data
| Related structure data |  6uqkMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_20849.map.gz / Format: CCP4 / Size: 149.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_20849.map.gz / Format: CCP4 / Size: 149.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | final map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.247 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
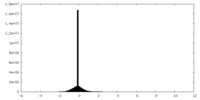
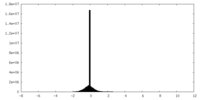
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_20849_msk_1.map emd_20849_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
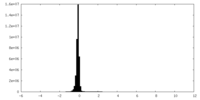
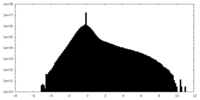
| Density Histograms |
-Half map: #1
| File | emd_20849_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
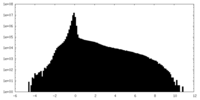
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_20849_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
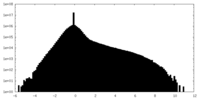
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : inositol 1,4,5-triphosphate receptor, type 3
| Entire | Name: inositol 1,4,5-triphosphate receptor, type 3 |
|---|---|
| Components |
|
-Supramolecule #1: inositol 1,4,5-triphosphate receptor, type 3
| Supramolecule | Name: inositol 1,4,5-triphosphate receptor, type 3 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 1.2 MDa |
-Macromolecule #1: inositol 1,4,5-triphosphate receptor, type 3
| Macromolecule | Name: inositol 1,4,5-triphosphate receptor, type 3 / type: protein_or_peptide / ID: 1 Details: The full sequence of the sample is MSSFLHIGDIVSLYAEGSVNGFISTLGLVDDRCVVEPAAGDLDNPPKKFRDCLFKVCPMNRYSAQKQYWKAKQTKQDKEK ...Details: The full sequence of the sample is MSSFLHIGDIVSLYAEGSVNGFISTLGLVDDRCVVEPAAGDLDNPPKKFRDCLFKVCPMNRYSAQKQYWKAKQTKQDKEK IADVVLLQKLQHAAQMEQKQNDTENKKVHGDVVKYGSVIQLLHMKSNKYLTVNKRLPALLEKNAMRVTLDATGNEGSWLF IQPFWKLRSNGDNVVVGDKVILNPVNAGQPLHASNYELSDNAGCKEVNSVNCNTSWKINLFMQFRDHLEEVLKGGDVVRL FHAEQEKFLTCDEYKGKLQVFLRTTLRQSATSATSSNALWEVEVVHHDPCRGGAGHWNGLYRFKHLATGNYLAAEENPSY KGDASDPKAAGMGAQGRTGRRNAGEKIKYCLVAVPHGNDIASLFELDPTTLQKTDSFVPRNSYVRLRHLCTNTWIQSTNV PIDIEEERPIRLMLGTCPTKEDKEAFAIVSVPVSEIRDLDFANDASSMLASAVEKLNEGFISQNDRRFVIQLLEDLVFFV SDVPNNGQNVLDIMVTKPNRERQKLMREQNILKQVFGILKAPFREKGGEGPLVRLEELSDQKNAPYQHMFRLCYRVLRHS QEDYRKNQEHIAKQFGMMQSQIGYDILAEDTITALLHNNRKLLEKHITKTEVETFVSLVRKNREPRFLDYLSDLCVSNHI AIPVTQELICKCVLDPKNSDILIRTELRPVKEMAQSHEYLSIEYSEEEVWLTWTDKNNEHHEKSVRQLAQEARAGNAHDE NVLSYYRYQLKLFARMCLDRQYLAIDEISQQLGVDLIFLCMADEMLPFDLRASFCHLMLHVHVDRDPQELVTPVKFARLW TEIPTAITIKDYDSNLNASRDDKKNKFANTMEFVEDYLNNVVSEAVPFANEEKNKLTFEVVSLAHNLIYFGFYSFSELLR LTRTLLGIIDCVQGPPAMLQAYEDPGGKNVRRSIQGVGHMMSTMVLSRKQSVFSAPSLSAGASAAEPLDRSKFEENEDIV VMETKLKILEILQFILNVRLDYRISYLLSVFKKEFVEVFPMQDSGADGTAPAFDSTTANMNLDRIGEQAEAMFGVGKTSS MLEVDDEGGRMFLRVLIHLTMHDYAPLVSGALQLLFKHFSQRQEAMHTFKQVQLLISAQDVENYKVIKSELDRLRTMVEK SELWVDKKGSGKGEEVEAGAAKDKKERPTDEEGFLHPPGEKSSENYQIVKGILERLNKMCGVGEQMRKKQQRLLKNMDAH KVMLDLLQIPYDKGDAKMMEILRYTHQFLQKFCAGNPGNQALLHKHLHLFLTPGLLEAETMQHIFLNNYQLCSEISEPVL QHFVHLLATHGRHVQYLDFLHTVIKAEGKYVKKCQDMIMTELTNAGDDVVVFYNDKASLAHLLDMMKAARDGVEDHSPLM YHISLVDLLAACAEGKNVYTEIKCTSLLPLEDVVSVVTHEDCITEVKMAYVNFVNHCYVDTEVEMKEIYTSNHIWTLFEN FTLDMARVCSKREKRVADPTLEKYVLSVVLDTINAFFSSPFSENSTSLQTHQTIVVQLLQSTTRLLECPWLQQQHKGSVE ACIRTLAMVAKGRAILLPMDLDAHISSMLSSGASCAAAAQRNASSYKATTRAFPRVTPTANQWDYKNIIEKLQDIITALE ERLKPLVQAELSVLVDVLHWPELLFLEGSEAYQRCESGGFLSKLIQHTKDLMESEEKLCIKVLRTLQQMLLKKTKYGDRG NQLRKMLLQNYLQNRKSTSRGDLPDPIGTGLDPDWSAIAATQCRLDKEGATKLVCDLITSTKNEKIFQESIGLAIHLLDG GNTEIQKSFHNLMMSDKKSERFFKVLHDRMKRAQQETKSTVAVNMNDLGSQPHEDREPVDPTTKGRVASFSIPGSSSRYS LGPSLRRGHEVSERVQSSEMGTSVLIMQPILRFLQLLCENHNRDLQNFLRCQNNKTNYNLVCETLQFLDIMCGSTTGGLG LLGLYINEDNVGLVIQTLETLTEYCQGPCHENQTCIVTHESNGIDIITALILNDISPLCKYRMDLVLQLKDNASKLLLAL MESRHDSENAERILISLRPQELVDVIKKAYLQEEERENSEVSPREVGHNIYILALQLSRHNKQLQHLLKPVKRIQEEEAE GISSMLSLNNKQLSQMLKSSAPAQEEEEDPLAYYENHTSQIEIVRQDRSMEQIVFPVPGICQFLTEETKHRLFTTTEQDE QGSKVSDFFDQSSFLHNEMEWQRKLRSMPLIYWFSRRMTLWGSISFNLAVFINIIIAFFYPYMEGASTGVLDSPLISLLF WILICFSIAALFTKRYSIRPLIVALILRSIYYLGIGPTLNILGALNLTNKIVFVVSFVGNRGTFIRGYKAMVMDMEFLYH VGYILTSVLGLFAHELFYSILLFDLIYREETLFNVIKSVTRNGRSILLTALLALILVYLFSIVGFLFLKDDFILEVDRLP NNHSTASPLGMPHGAAAFVDTCSGDKMDCVSGLSVPEVLEEDRELDSTERACDTLLMCIVTVMNHGLRNGGGVGDILRKP SKDESLFPARVVYDLLFFFIVIIIVLNLIFGVIIDTFADLRSEKQKKEEILKTTCFICGLERDKFDNKTVSFEEHIKLEH NMWNYLYFIVLVRVKNKTDYTGPESYVAQMIKNKNLDWFPRMRAMSLVSNEGEGEQNEIRILQDKLNSTMKLVSHLTAQL NELKEQMTEQRKRRQRLGFVDVQNCISRGENLYFQSAWSHPQFEKGGGSGGGSGGSAWSHPQFEK Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 278.584406 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSSFLHIGDI VSLYAEGSVN GFISTLGLVD DRCVVEPAAG DLDNPPKKFR DCLFKVCPMN RYSAQKQYWK AKQTKQDKEK IADVVLLQK LQHAAQMEQK QNDTENKKVH GDVVKYGSVI QLLHMKSNKY LTVNKRLPAL LEKNAMRVTL DATGNEGSWL F IQPFWKLR ...String: MSSFLHIGDI VSLYAEGSVN GFISTLGLVD DRCVVEPAAG DLDNPPKKFR DCLFKVCPMN RYSAQKQYWK AKQTKQDKEK IADVVLLQK LQHAAQMEQK QNDTENKKVH GDVVKYGSVI QLLHMKSNKY LTVNKRLPAL LEKNAMRVTL DATGNEGSWL F IQPFWKLR SNGDNVVVGD KVILNPVNAG QPLHASNYEL SDNAGCKEVN SVNCNTSWKI NLFMQFRDHL EEVLKGGDVV RL FHAEQEK FLTCDEYKGK LQVFLRTTLR QSATSATSSN ALWEVEVVHH DPCRGGAGHW NGLYRFKHLA TGNYLAAEEN PSY KGDASD PKAAGMGAQG RTGRRNAGEK IKYCLVAVPH GNDIASLFEL DPTTLQKTDS FVPRNSYVRL RHLCTNTWIQ STNV PIDIE EERPIRLMLG TCPTKEDKEA FAIVSVPVSE IRDLDFANDA SSMLASAVEK LNEGFISQND RRFVIQLLED LVFFV SDVP NNGQNVLDIM VTKPNRERQK LMREQNILKQ VFGILKAPFR EKGGEGPLVR LEELSDQKNA PYQHMFRLCY RVLRHS QED YRKNQEHIAK QFGMMQSQIG YDILAEDTIT ALLHNNRKLL EKHITKTEVE TFVSLVRKNR EPRFLDYLSD LCVSNHI AI PVTQELICKC VLDPKNSDIL IRTELRPVKE MAQSHEYLSI EYSEEEVWLT WTDKNNEHHE KSVRQLAQEA RAGNAHDE N VLSYYRYQLK LFARMCLDRQ YLAIDEISQQ LGVDLIFLCM ADEMLPFDLR ASFCHLMLHV HVDRDPQELV TPVKFARLW TEIPTAITIK DYDSNLNASR DDKKNKFANT MEFVEDYLNN VVSEAVPFAN EEKNKLTFEV VSLAHNLIYF GFYSFSELLR LTRTLLGII DCVQGPPAML QAYEDPGGKN VRRSIQGVGH MMSTMVLSRK QSVFSAPSLS AGASAAEPLD RSKFEENEDI V VMETKLKI LEILQFILNV RLDYRISYLL SVFKKEFVEV FPMQDSGADG TAPAFDSTTA NMNLDRIGEQ AEAMFGVGKT SS MLEVDDE GGRMFLRVLI HLTMHDYAPL VSGALQLLFK HFSQRQEAMH TFKQVQLLIS AQDVENYKVI KSELDRLRTM VEK SELWVD KKGSGKGEEV EAGAAKDKKE RPTDEEGFLH PPGEKSSENY QIVKGILERL NKMCGVGEQM RKKQQRLLKN MDAH KVMLD LLQIPYDKGD AKMMEILRYT HQFLQKFCAG NPGNQALLHK HLHLFLTPGL LEAETMQHIF LNNYQLCSEI SEPVL QHFV HLLATHGRHV QYLDFLHTVI KAEGKYVKKC QDMIMTELTN AGDDVVVFYN DKASLAHLLD MMKAARDGVE DHSPLM YHI SLVDLLAACA EGKNVYTEIK CTSLLPLEDV VSVVTHEDCI TEVKMAYVNF VNHC(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)DYKNIIEKL QDIITALEER L KPLVQAEL SVLVDVLHWP ELLFLEGSEA YQRCESGGFL SKLIQHTKDL MESEEKLCIK VLRTLQQMLL KKTKYGDRGN QL RKMLLQN YLQNRKSTSR GDLPDPIGTG LDPDWSAIAA TQCRLDKEGA TKLVCDLITS TKNEKIFQES IGLAIHLLDG GNT EIQKSF HNLMMSDKKS ERFFKVLHDR MKRAQQETKS TVAVNMNDLG SQPHEDREPV DPTTKGRVAS FSIPGSSSRY SLGP SLRRG HEVSERVQSS EMGTSVLIMQ PILRFLQLLC ENHNRDLQNF LRCQNNKTNY NLVCETLQFL DIMCGSTTGG LGLLG LYIN EDNVGLVIQT LETLTEYCQG PCHENQTCIV THESNGIDII TALILNDISP LCKYRMDLVL QLKDNASKLL LALMES RHD SENAERILIS LRPQELVDVI KKAYLQEEER ENSEVSPREV GHNIYILALQ LSRHNKQLQH LLKPVKRIQE EEAEGIS SM LSLNNKQLSQ MLKSSAPAQE EEEDPLAYYE NHTSQIEIVR QDRSMEQIVF PVPGICQFLT EETKHRLFTT TEQDEQGS K VSDFFDQSSF LHNEMEWQRK LRSMPLIYWF SRRMTLWGSI SFNLAVFINI IIAFFYPYME (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) PLIVALILRS IYYLGIGPTL NILGALNLTN KIVFVVSFVG NRGTFIRGYK AMVMDMEFL YHVGYILTSV LGLFAHELFY SILLFDLIYR EETLFNVIKS VTRNGRSILL TALLALILVY LFSIVGFLFL K DDFILEVD RLPNNHSTAS PLGMPHGAAA FVDTCSGDKM DCVSGLSVPE VLEEDRELDS TERACDTLLM CIVTVMNHGL RN GGGVGDI LRKPSKDESL FPARVVYDLL FFFIVIIIVL NLIFGVIIDT FADLRSEKQK KEEILKTTCF ICGLERDKFD NKT VSFEEH IKLEHNMWNY LYFIVLVRVK NKTDYTGPES YVAQMIKNKN LDWFPRMRAM SLV(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK) |
-Macromolecule #2: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 2 / Number of copies: 4 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.3 mg/mL | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| ||||||||||||||
| Grid | Model: C-flat-2/1 / Material: COPPER / Mesh: 200 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 120 sec. / Details: 25 mA | ||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 281 K / Instrument: FEI VITROBOT MARK IV / Details: The grid was blotted for 3 seconds at force 1. |
- Electron microscopy
Electron microscopy
| Microscope | FEI POLARA 300 |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 70.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.2 mm / Nominal magnification: 31000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: EMDB MAP EMDB ID: |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C4 (4 fold cyclic) / Resolution.type: BY AUTHOR / Resolution: 3.77 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: cisTEM / Number images used: 82511 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller


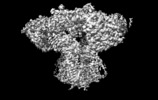





























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)