+Search query
-Structure paper
| Title | Cryo-EM reveals ligand induced allostery underlying InsPR channel gating. |
|---|---|
| Journal, issue, pages | Cell Res, Vol. 28, Issue 12, Page 1158-1170, Year 2018 |
| Publish date | Nov 23, 2018 |
 Authors Authors | Guizhen Fan / Mariah R Baker / Zhao Wang / Alexander B Seryshev / Steven J Ludtke / Matthew L Baker / Irina I Serysheva /  |
| PubMed Abstract | Inositol-1,4,5-trisphosphate receptors (InsPRs) are cation channels that mobilize Ca from intracellular stores in response to a wide range of cellular stimuli. The paradigm of InsPR activation is the ...Inositol-1,4,5-trisphosphate receptors (InsPRs) are cation channels that mobilize Ca from intracellular stores in response to a wide range of cellular stimuli. The paradigm of InsPR activation is the coupled interplay between binding of InsP and Ca that switches the ion conduction pathway between closed and open states to enable the passage of Ca through the channel. However, the molecular mechanism of how the receptor senses and decodes ligand-binding signals into gating motion remains unknown. Here, we present the electron cryo-microscopy structure of InsPR1 from rat cerebellum determined to 4.1 Å resolution in the presence of activating concentrations of Ca and adenophostin A (AdA), a structural mimetic of InsP and the most potent known agonist of the channel. Comparison with the 3.9 Å-resolution structure of InsPR1 in the Apo-state, also reported herein, reveals the binding arrangement of AdA in the tetrameric channel assembly and striking ligand-induced conformational rearrangements within cytoplasmic domains coupled to the dilation of a hydrophobic constriction at the gate. Together, our results provide critical insights into the mechanistic principles by which ligand-binding allosterically gates InsPR channel. |
 External links External links |  Cell Res / Cell Res /  PubMed:30470765 / PubMed:30470765 /  PubMed Central PubMed Central |
| Methods | EM (single particle) |
| Resolution | 3.9 - 4.3 Å |
| Structure data | EMDB-9243: Structure of full-length IP3R1 channel bound with Adenophostin A (composite) EMDB-9244: Structure of full-length IP3R1 channel in Apo-state (composite)  EMDB-9245:  EMDB-9246:  EMDB-9247:  EMDB-9248: |
| Chemicals | 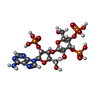 ChemComp-JYP: |
| Source |
|
 Keywords Keywords | MEMBRANE PROTEIN / inositol 1 / 4 / 5-trisphosphate receptor / calcium release channel / neuronal type 1 / adenophostin A / IP3R-channel activator |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers








