[English] 日本語
 Yorodumi
Yorodumi- EMDB-8120: Structure of TRPV1 in complex with capsazepine, determined in lip... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-8120 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of TRPV1 in complex with capsazepine, determined in lipid nanodisc | |||||||||||||||
 Map data Map data | None | |||||||||||||||
 Sample Sample |
| |||||||||||||||
| Biological species |  | |||||||||||||||
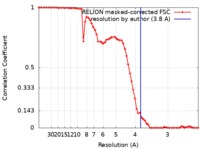
| Method | single particle reconstruction / cryo EM / Resolution: 3.8 Å | |||||||||||||||
 Authors Authors | Gao Y / Cao E / Julius D / Cheng Y | |||||||||||||||
| Funding support |  United States, 4 items United States, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Nature / Year: 2016 Journal: Nature / Year: 2016Title: TRPV1 structures in nanodiscs reveal mechanisms of ligand and lipid action. Authors: Yuan Gao / Erhu Cao / David Julius / Yifan Cheng /  Abstract: When integral membrane proteins are visualized in detergents or other artificial systems, an important layer of information is lost regarding lipid interactions and their effects on protein structure. ...When integral membrane proteins are visualized in detergents or other artificial systems, an important layer of information is lost regarding lipid interactions and their effects on protein structure. This is especially relevant to proteins for which lipids have both structural and regulatory roles. Here we demonstrate the power of combining electron cryo-microscopy with lipid nanodisc technology to ascertain the structure of the rat TRPV1 ion channel in a native bilayer environment. Using this approach, we determined the locations of annular and regulatory lipids and showed that specific phospholipid interactions enhance binding of a spider toxin to TRPV1 through formation of a tripartite complex. Furthermore, phosphatidylinositol lipids occupy the binding site for capsaicin and other vanilloid ligands, suggesting a mechanism whereby chemical or thermal stimuli elicit channel activation by promoting the release of bioactive lipids from a critical allosteric regulatory site. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_8120.map.gz emd_8120.map.gz | 25 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-8120-v30.xml emd-8120-v30.xml emd-8120.xml emd-8120.xml | 21.5 KB 21.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_8120_fsc.xml emd_8120_fsc.xml | 6.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_8120.png emd_8120.png | 87.7 KB | ||
| Others |  emd_8120_half_map_1.map.gz emd_8120_half_map_1.map.gz emd_8120_half_map_2.map.gz emd_8120_half_map_2.map.gz | 17.8 MB 17.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-8120 http://ftp.pdbj.org/pub/emdb/structures/EMD-8120 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8120 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8120 | HTTPS FTP |
-Related structure data
| Related structure data |  8117C  8118C  8119C  5irxC  5irzC  5is0C C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_8120.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_8120.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | None | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.2156 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
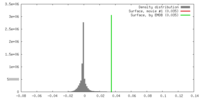
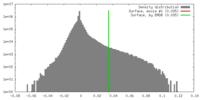
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: None
| File | emd_8120_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | None | ||||||||||||
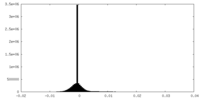
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: None
| File | emd_8120_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | None | ||||||||||||
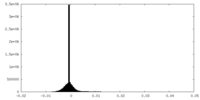
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : TRPV1 ion channel in complex with capsazepine
| Entire | Name: TRPV1 ion channel in complex with capsazepine |
|---|---|
| Components |
|
-Supramolecule #1: TRPV1 ion channel in complex with capsazepine
| Supramolecule | Name: TRPV1 ion channel in complex with capsazepine / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  Homo sapiens (human) / Recombinant cell: HEK 293S Homo sapiens (human) / Recombinant cell: HEK 293S |
-Macromolecule #1: The capsaicin receptor, TRPV1
| Macromolecule | Name: The capsaicin receptor, TRPV1 / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: AMGSRLYDRR SIFDAVAQSN CQELESLLPF LQRSKKRLTD SEFKDPETGK TCLLKAMLNL HNGQNDTIAL LLDVARKTDS LKQFVNASYT DSYYKGQTAL HIAIERRNMT LVTLLVENGA DVQAAANGDF FKKTKGRPGF YFGELPLSLA ACTNQLAIVK FLLQNSWQPA ...String: AMGSRLYDRR SIFDAVAQSN CQELESLLPF LQRSKKRLTD SEFKDPETGK TCLLKAMLNL HNGQNDTIAL LLDVARKTDS LKQFVNASYT DSYYKGQTAL HIAIERRNMT LVTLLVENGA DVQAAANGDF FKKTKGRPGF YFGELPLSLA ACTNQLAIVK FLLQNSWQPA DISARDSVGN TVLHALVEVA DNTVDNTKFV TSMYNEILIL GAKLHPTLKL EEITNRKGLT PLALAASSGK IGVLAYILQR EIHEPECRHL SRKFTEWAYG PVHSSLYDLS CIDTCEKNSV LEVIAYSSSE TPNRHDMLLV EPLNRLLQDK WDRFVKRIFY FNFFVYCLYM IIFTAAAYYR PVEGLPPYKL KNTVGDYFRV TGEILSVSGG VYFFFRGIQY FLQRRPSLKS LFVDSYSEIL FFVQSLFMLV SVVLYFSQRK EYVASMVFSL AMGWTNMLYY TRGFQQMGIY AVMIEKMILR DLCRFMFVYL VFLFGFSTAV VTLIEDGKYN SLYSTCLELF KFTIGMGDLE FTENYDFKAV FIILLLAYVI LTYILLLNML IALMGETVNK IAQESKNIWK LQRAITILDT EKSFLKCMRK AFRSGKLLQV GFTPDGKDDY RWCFRVDEVN WTTWNTNVGI INEDPG |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.5 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
| ||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: HOLEY / Support film - Film thickness: 12.0 nm / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 290.0 kPa | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 298 K / Instrument: FEI VITROBOT MARK III Details: Blot for 6 seconds before plunging into liquid ethane (FEI VITROBOT MARK III).. | ||||||||||||
| Details | This sample was monodisperse. |
- Electron microscopy
Electron microscopy
| Microscope | FEI POLARA 300 |
|---|---|
| Temperature | Min: 70.0 K / Max: 70.0 K |
| Details | Grid screening was performed manually. |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Digitization - Dimensions - Width: 3838 pixel / Digitization - Dimensions - Height: 3710 pixel / Digitization - Frames/image: 1-30 / Number grids imaged: 1 / Number real images: 1002 / Average exposure time: 6.0 sec. / Average electron dose: 41.0 e/Å2 Details: Images were collected in movie mode at 5 frames per second for 6 seconds. |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 30.0 µm / Calibrated defocus max: 2.2 µm / Calibrated defocus min: 0.7 µm / Calibrated magnification: 41132 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.0 mm / Nominal defocus max: 2.2 µm / Nominal defocus min: 0.7 µm / Nominal magnification: 31000 |
| Sample stage | Specimen holder model: OTHER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Chain ID: A |
|---|---|
| Refinement | Space: REAL / Protocol: RIGID BODY FIT / Target criteria: correlation coefficient |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)