[English] 日本語
 Yorodumi
Yorodumi- EMDB-20145: Cryo-EM structure of the C2-symmetric TRPV2/RTx complex in amphip... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-20145 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the C2-symmetric TRPV2/RTx complex in amphipol resolved to 3.3 A | |||||||||
 Map data Map data | C2-symmetric TRPV2/RTx complex in amphipol resolved to 3.3 A | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | ion channel / calcium channel / TRP channel / metal transport | |||||||||
| Function / homology |  Function and homology information Function and homology informationgrowth cone membrane / response to temperature stimulus / positive regulation of calcium ion import / calcium ion import across plasma membrane / positive regulation of axon extension / axonal growth cone / calcium channel activity / positive regulation of cold-induced thermogenesis / cell body / cell surface / identical protein binding Similarity search - Function | |||||||||
| Biological species |  | |||||||||
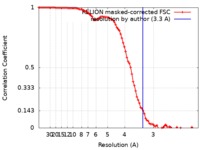
| Method | single particle reconstruction / cryo EM / Resolution: 3.3 Å | |||||||||
 Authors Authors | Zubcevic L / Hsu AL | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Elife / Year: 2019 Journal: Elife / Year: 2019Title: Symmetry transitions during gating of the TRPV2 ion channel in lipid membranes. Authors: Lejla Zubcevic / Allen L Hsu / Mario J Borgnia / Seok-Yong Lee /  Abstract: The Transient Receptor Potential Vanilloid 2 (TRPV2) channel is a member of the temperature-sensing thermoTRPV family. Recent advances in cryo-electronmicroscopy (cryo-EM) and X-ray crystallography ...The Transient Receptor Potential Vanilloid 2 (TRPV2) channel is a member of the temperature-sensing thermoTRPV family. Recent advances in cryo-electronmicroscopy (cryo-EM) and X-ray crystallography have provided many important insights into the gating mechanisms of thermoTRPV channels. Interestingly, crystallographic studies of ligand-dependent TRPV2 gating have shown that the TRPV2 channel adopts two-fold symmetric arrangements during the gating cycle. However, it was unclear if crystal packing forces played a role in stabilizing the two-fold symmetric arrangement of the channel. Here, we employ cryo-EM to elucidate the structure of full-length rabbit TRPV2 in complex with the agonist resiniferatoxin (RTx) in nanodiscs and amphipol. We show that RTx induces two-fold symmetric conformations of TRPV2 in both environments. However, the two-fold symmetry is more pronounced in the native-like lipid environment of the nanodiscs. Our data offers insights into a gating pathway in TRPV2 involving symmetry transitions. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_20145.map.gz emd_20145.map.gz | 59.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-20145-v30.xml emd-20145-v30.xml emd-20145.xml emd-20145.xml | 18.6 KB 18.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_20145_fsc.xml emd_20145_fsc.xml | 9.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_20145.png emd_20145.png | 84.4 KB | ||
| Filedesc metadata |  emd-20145.cif.gz emd-20145.cif.gz | 6.5 KB | ||
| Others |  emd_20145_half_map_1.map.gz emd_20145_half_map_1.map.gz emd_20145_half_map_2.map.gz emd_20145_half_map_2.map.gz | 48.5 MB 48.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-20145 http://ftp.pdbj.org/pub/emdb/structures/EMD-20145 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20145 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20145 | HTTPS FTP |
-Related structure data
| Related structure data |  6oo4MC  6oo3C  6oo5C  6oo7C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_20145.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_20145.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | C2-symmetric TRPV2/RTx complex in amphipol resolved to 3.3 A | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.08 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: Half map 1
| File | emd_20145_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map 2
| File | emd_20145_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : TRPV2
| Entire | Name: TRPV2 |
|---|---|
| Components |
|
-Supramolecule #1: TRPV2
| Supramolecule | Name: TRPV2 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 Details: TRPV2 in complex with RTx reconstituted into amphipol A8-35 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 300 KDa |
-Macromolecule #1: TRPV2
| Macromolecule | Name: TRPV2 / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 88.72268 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MTSPSSPPAF RLETSDGGQD GAEVDKAQLG YGAGPPPMES RFQDEDRNFP PQIKVNLNYR KGAGASQPDL NRFDRDRLFN VVARGNPED LAGLLEYLRR TSKYLTDSEY TEGSTGKTCL MKAVLNLQDG VNACIQPLLE IDRDSGNPQP LVNAQCTDEY Y RGHSALHI ...String: MTSPSSPPAF RLETSDGGQD GAEVDKAQLG YGAGPPPMES RFQDEDRNFP PQIKVNLNYR KGAGASQPDL NRFDRDRLFN VVARGNPED LAGLLEYLRR TSKYLTDSEY TEGSTGKTCL MKAVLNLQDG VNACIQPLLE IDRDSGNPQP LVNAQCTDEY Y RGHSALHI AIEKRSLQCV KLLVENGANV HAKACGHFFQ KNQDTCFYFG ELPLSLAACT KQWDVVNYLL ENPHQPASLQ AQ DSLGNTV LHALVMIADD SAENSALVVR MYDGLLQAGA RLCPNVQLEG IPNLEGLTPL KLAAKEGKIE IFKHILQREF SAP CQSLSR KFTEWCYGPV RVSLYDLASV DSWEENSVLE IIAFHSRSPH RHRMVVLEPL NKLLQAKWDR LIPRFCFNFL CYLV YMLIF TAVAYHQPAL EKQGFPPLKA TAGNSMLLLG HILILLGGVY LLLGQLWYFW RRRLFIWISF MDSYSEILFL LQALL TVLS QVLCFLAIEW YLPLLVSSLA MGWTNLLYYT RGFQHTGIYS VMIEKVILRD LLRFLLVYLV FLFGFAVALV SLSREA QNS RTPAGPNATE VGQPGAGQED EAPPYRSILD ASLELFKFTI GMGELAFQEQ LRFRGVVLLL LLAYVLLTYV LLLNMLI AL MSETVNSVAT DSWSIWKLQK AISVLEMENG YWWCRRKKQR AGVMLTVGTR PDGSPDERWC FRVGEMNWAT WEQTLPRT L CEEPSGAAAP GVMKNPTPAS QRGEDSASEE DHLPLQLLQS RSNLEVLFQG PHHHHHHDYK DDDDK UniProtKB: Transient receptor potential cation channel subfamily V member 2 |
-Macromolecule #2: resiniferatoxin
| Macromolecule | Name: resiniferatoxin / type: ligand / ID: 2 / Number of copies: 4 / Formula: 6EU |
|---|---|
| Molecular weight | Theoretical: 628.708 Da |
| Chemical component information | 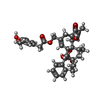 ChemComp-6EU: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| ||||||||||||
| Grid | Model: Quantifoil, UltrAuFoil, R1.2/1.3 / Material: GOLD / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 296 K / Instrument: LEICA EM GP / Details: Blotted 3 seconds before plunging. | ||||||||||||
| Details | TRPV2 in complex with RTx reconstituted into amphipol A8-35, monodisperse |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Number real images: 1293 / Average electron dose: 42.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source: OTHER |
| Electron optics | Illumination mode: OTHER / Imaging mode: OTHER |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller


 UCSF Chimera
UCSF Chimera











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)