+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-23493 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of squirrel TRPV1 in complex with capsaicin | |||||||||||||||
 Map data Map data | ||||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | Transient Receptor Potential V Family Member 1 / TRP channel / TRPV1 full length / TRPV1 wild type / capsaicin / MEMBRANE PROTEIN | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationresponse to capsazepine / sensory perception of mechanical stimulus / peptide secretion / smooth muscle contraction involved in micturition / temperature-gated ion channel activity / detection of chemical stimulus involved in sensory perception of pain / fever generation / detection of temperature stimulus involved in thermoception / cellular response to acidic pH / dendritic spine membrane ...response to capsazepine / sensory perception of mechanical stimulus / peptide secretion / smooth muscle contraction involved in micturition / temperature-gated ion channel activity / detection of chemical stimulus involved in sensory perception of pain / fever generation / detection of temperature stimulus involved in thermoception / cellular response to acidic pH / dendritic spine membrane / diet induced thermogenesis / detection of temperature stimulus involved in sensory perception of pain / behavioral response to pain / calcium ion import across plasma membrane / extracellular ligand-gated monoatomic ion channel activity / phosphoprotein binding / GABA-ergic synapse / lipid metabolic process / calcium channel activity / sensory perception of taste / cellular response to heat / postsynaptic membrane / calmodulin binding / negative regulation of transcription by RNA polymerase II / ATP binding / metal ion binding Similarity search - Function | |||||||||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  Ictidomys tridecemlineatus (thirteen-lined ground squirrel) Ictidomys tridecemlineatus (thirteen-lined ground squirrel) | |||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.81 Å | |||||||||||||||
 Authors Authors | Neuberger A / Nadezhdin KD | |||||||||||||||
| Funding support |  United States, 4 items United States, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2021 Journal: Nat Commun / Year: 2021Title: Extracellular cap domain is an essential component of the TRPV1 gating mechanism. Authors: Kirill D Nadezhdin / Arthur Neuberger / Yury A Nikolaev / Lyle A Murphy / Elena O Gracheva / Sviatoslav N Bagriantsev / Alexander I Sobolevsky /  Abstract: Transient receptor potential (TRP) channels are polymodal molecular sensors involved in numerous physiological processes and implicated in a variety of human diseases. Several structures of the ...Transient receptor potential (TRP) channels are polymodal molecular sensors involved in numerous physiological processes and implicated in a variety of human diseases. Several structures of the founding member of the TRP channel family, TRPV1, are available, all of which were determined for the protein missing the N- and C-termini and the extracellular S5-P-loop. Here, we present structures of the full-length thirteen-lined ground squirrel TRPV1 solved by cryo-EM. Our structures resolve the extracellular cap domain formed by the S5-P-loops and the C-terminus that wraps around the three-stranded β-sheet connecting elements of the TRPV1 intracellular skirt. The cap domain forms a dome above the pore's extracellular entrance, with four portals leading to the ion conductance pathway. Deletion of the cap increases the TRPV1 average conductance, reduces the open probability and affects ion selectivity. Our data show that both the termini and the cap domain are critical determinants of TRPV1 function. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_23493.map.gz emd_23493.map.gz | 59.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-23493-v30.xml emd-23493-v30.xml emd-23493.xml emd-23493.xml | 15.7 KB 15.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_23493.png emd_23493.png | 182.5 KB | ||
| Filedesc metadata |  emd-23493.cif.gz emd-23493.cif.gz | 6.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-23493 http://ftp.pdbj.org/pub/emdb/structures/EMD-23493 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23493 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23493 | HTTPS FTP |
-Related structure data
| Related structure data |  7lr0MC  7lqyC  7lqzC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_23493.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_23493.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.83 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : sample 1
| Entire | Name: sample 1 |
|---|---|
| Components |
|
-Supramolecule #1: sample 1
| Supramolecule | Name: sample 1 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Osm-9-like TRP channel 1
| Macromolecule | Name: Osm-9-like TRP channel 1 / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Ictidomys tridecemlineatus (thirteen-lined ground squirrel) Ictidomys tridecemlineatus (thirteen-lined ground squirrel) |
| Molecular weight | Theoretical: 95.902781 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MKKWANLDSG ESEDPPHEDS CPDPPDGDPN SKQSPAKPHI FSTTKSRTRL FGKGDSEEAS PMDCSYEEGE LASCPTITVS SVVIIQRPG DALTCARQLS QDSVSAGAEK PLKLYDRRSI FDAVAQNNCQ ELESLLPFLQ KSRKRLTDSE FKDPETGKTC L LKAMLNLH ...String: MKKWANLDSG ESEDPPHEDS CPDPPDGDPN SKQSPAKPHI FSTTKSRTRL FGKGDSEEAS PMDCSYEEGE LASCPTITVS SVVIIQRPG DALTCARQLS QDSVSAGAEK PLKLYDRRSI FDAVAQNNCQ ELESLLPFLQ KSRKRLTDSE FKDPETGKTC L LKAMLNLH NGQNDTISLL LDIARQTDSL KEFVNASYTD SYYKGQTALH IAIERRNMAL VTLLVENGAD VQAAANGDFF KK TKGRPGF YFGELPLSLA ACTNQLAIVK FLLQNSWQPA DISARDSVGN TVLHALVEVA DNTADNTKFV TSMYNEILIL GAK LHPTLK LEELINKKGL TPLALAASSG KIGVLAYILQ REIQEPECRH LSRKFTEWAY GPVHSSLYDL SCIDTCEKNS VLEV IAYSS SETPNRHDML LVEPLNRLLQ DKWDRFVKRI FYFNFFVYCL YMIVFTTAAY YRPVDGLPPY KLKNTVGDYF RVTGE ILSV SGGVYFFLRG IQYFLQRRPS MKTLFVDSYS EMLFFVQSLF MLGSVVLYFS HRKEYVASMV FSLAMGWTNM LYYTRG FQQ MGIYAVMIEK MILRDLCRFM FVYLVFLFGF STAVVTLIED GKNYSEPAEY TSHRWRAPGC RPPDSYNSLY STCLELF KF TIGMGDLEFT ENYDFKAVFI ILLLAYVILT YILLLNMLIA LMGETVNKIA QESKNIWKLQ RAITILDTEK SFLKCMRK A FRSGKLLQVG YTPDGKDDYR WCFRVDEVNW TTWNTNVGII NEDPGNCEGV KRTLSFSLRS GRVSGRNWRN FALVPLLRD ASTRDRHHAQ PEEVHLKHFA GSLKPEDAEV FKDSMAPGEK LPVR UniProtKB: Transient receptor potential cation channel subfamily V member 1 |
-Macromolecule #2: (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(tri...
| Macromolecule | Name: (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate type: ligand / ID: 2 / Number of copies: 28 / Formula: POV |
|---|---|
| Molecular weight | Theoretical: 760.076 Da |
| Chemical component information |  ChemComp-POV: |
-Macromolecule #3: (6E)-N-(4-hydroxy-3-methoxybenzyl)-8-methylnon-6-enamide
| Macromolecule | Name: (6E)-N-(4-hydroxy-3-methoxybenzyl)-8-methylnon-6-enamide type: ligand / ID: 3 / Number of copies: 4 / Formula: 4DY |
|---|---|
| Molecular weight | Theoretical: 305.412 Da |
| Chemical component information | 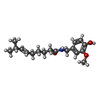 ChemComp-4DY: |
-Macromolecule #4: SODIUM ION
| Macromolecule | Name: SODIUM ION / type: ligand / ID: 4 / Number of copies: 1 |
|---|---|
| Molecular weight | Theoretical: 22.99 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2.4 mg/mL | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| ||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Specialist optics | Energy filter - Slit width: 30 eV |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Digitization - Dimensions - Width: 5760 pixel / Digitization - Dimensions - Height: 4092 pixel / Number grids imaged: 1 / Number real images: 9453 / Average exposure time: 2.5 sec. / Average electron dose: 58.5 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: -2.5 µm / Nominal defocus min: -1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL |
|---|---|
| Output model |  PDB-7lr0: |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















