[English] 日本語
 Yorodumi
Yorodumi- EMDB-7896: HIV Env BG505 SOSIP with polyclonal Fabs from immunized rabbit #3... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-7896 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | HIV Env BG505 SOSIP with polyclonal Fabs from immunized rabbit #3417 post-boost#1 | |||||||||
 Map data Map data | HIV Env BG505 SOSIP with polyclonal Fabs from immunized rabbit #3417 post-boost#1 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | HIV / antibodies / antibody / VIRAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont-mediated perturbation of host defense response / immunoglobulin complex / positive regulation of plasma membrane raft polarization / positive regulation of receptor clustering / host cell endosome membrane / clathrin-dependent endocytosis of virus by host cell / adaptive immune response / viral protein processing / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane ...symbiont-mediated perturbation of host defense response / immunoglobulin complex / positive regulation of plasma membrane raft polarization / positive regulation of receptor clustering / host cell endosome membrane / clathrin-dependent endocytosis of virus by host cell / adaptive immune response / viral protein processing / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / virion attachment to host cell / host cell plasma membrane / virion membrane / structural molecule activity / extracellular region / identical protein binding / membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /   Human immunodeficiency virus 1 / Human immunodeficiency virus 1 /  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.71 Å | |||||||||
 Authors Authors | Turner HL / Cottrell CA | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Immunity / Year: 2018 Journal: Immunity / Year: 2018Title: Electron-Microscopy-Based Epitope Mapping Defines Specificities of Polyclonal Antibodies Elicited during HIV-1 BG505 Envelope Trimer Immunization. Authors: Matteo Bianchi / Hannah L Turner / Bartek Nogal / Christopher A Cottrell / David Oyen / Matthias Pauthner / Raiza Bastidas / Rebecca Nedellec / Laura E McCoy / Ian A Wilson / Dennis R Burton ...Authors: Matteo Bianchi / Hannah L Turner / Bartek Nogal / Christopher A Cottrell / David Oyen / Matthias Pauthner / Raiza Bastidas / Rebecca Nedellec / Laura E McCoy / Ian A Wilson / Dennis R Burton / Andrew B Ward / Lars Hangartner /   Abstract: Characterizing polyclonal antibody responses via currently available methods is inherently complex and difficult. Mapping epitopes in an immune response is typically incomplete, which creates a ...Characterizing polyclonal antibody responses via currently available methods is inherently complex and difficult. Mapping epitopes in an immune response is typically incomplete, which creates a barrier to fully understanding the humoral response to antigens and hinders rational vaccine design efforts. Here, we describe a method of characterizing polyclonal responses by using electron microscopy, and we applied this method to the immunization of rabbits with an HIV-1 envelope glycoprotein vaccine candidate, BG505 SOSIP.664. We detected known epitopes within the polyclonal sera and revealed how antibody responses evolved during the prime-boosting strategy to ultimately result in a neutralizing antibody response. We uncovered previously unidentified epitopes, including an epitope proximal to one recognized by human broadly neutralizing antibodies as well as potentially distracting non-neutralizing epitopes. Our method provides an efficient and semiquantitative map of epitopes that are targeted in a polyclonal antibody response and should be of widespread utility in vaccine and infection studies. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_7896.map.gz emd_7896.map.gz | 97.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-7896-v30.xml emd-7896-v30.xml emd-7896.xml emd-7896.xml | 17.3 KB 17.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_7896.png emd_7896.png | 22 KB | ||
| Filedesc metadata |  emd-7896.cif.gz emd-7896.cif.gz | 6.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-7896 http://ftp.pdbj.org/pub/emdb/structures/EMD-7896 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7896 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7896 | HTTPS FTP |
-Related structure data
| Related structure data |  6didMC  7552C  7553C  7554C  7555C  7556C  7557C  7570C 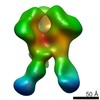 7887C 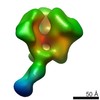 7888C 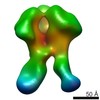 7889C 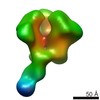 7890C 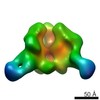 7891C 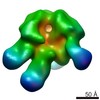 7892C 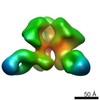 7893C  7894C  7895C  7903C  7904C  7906C  6cjkC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_7896.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_7896.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | HIV Env BG505 SOSIP with polyclonal Fabs from immunized rabbit #3417 post-boost#1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.15 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : HIV Env BG505 SOSIP with polyclonal Fabs from immunized rabbit #3...
| Entire | Name: HIV Env BG505 SOSIP with polyclonal Fabs from immunized rabbit #3417 post-boost#1 |
|---|---|
| Components |
|
-Supramolecule #1: HIV Env BG505 SOSIP with polyclonal Fabs from immunized rabbit #3...
| Supramolecule | Name: HIV Env BG505 SOSIP with polyclonal Fabs from immunized rabbit #3417 post-boost#1 type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 Details: Model includes fitted atomic structures for HIV Env BG505 SOSIP (PDB ID: 5v8m) and 10A Fab (PDB ID: 6cjk). |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Envelope glycoprotein gp160
| Macromolecule | Name: Envelope glycoprotein gp160 / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Molecular weight | Theoretical: 54.064277 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: AENLWVTVYY GVPVWKDAET TLFCASDAKA YETEKHNVWA THACVPTDPN PQEIHLENVT EEFNMWKNNM VEQMHTDIIS LWDQSLKPC VKLTPLCVTL QCTNVTNNIT DDMRGELKNC SFNMTTELRD KKQKVYSLFY RLDVVQINEN QGNRSNNSNK E YRLINCNT ...String: AENLWVTVYY GVPVWKDAET TLFCASDAKA YETEKHNVWA THACVPTDPN PQEIHLENVT EEFNMWKNNM VEQMHTDIIS LWDQSLKPC VKLTPLCVTL QCTNVTNNIT DDMRGELKNC SFNMTTELRD KKQKVYSLFY RLDVVQINEN QGNRSNNSNK E YRLINCNT SAITQACPKV SFEPIPIHYC APAGFAILKC KDKKFNGTGP CPSVSTVQCT HGIKPVVSTQ LLLNGSLAEE EV MIRSENI TNNAKNILVQ FNTPVQINCT RPNNNTRKSI RIGPGQAFYA TGDIIGDIRQ AHCNVSKATW NETLGKVVKQ LRK HFGNNT IIRFANSSGG DLEVTTHSFN CGGEFFYCNT SGLFNSTWIS NTSVQGSNST GSNDSITLPC RIKQIINMWQ RIGQ AMYAP PIQGVIRCVS NITGLILTRD GGSTNSTTET FRPGGGDMRD NWRSELYKYK VVKIEPLGVA PTRCKRRVVG RRRRR R UniProtKB: Envelope glycoprotein gp160 |
-Macromolecule #2: Envelope glycoprotein gp160
| Macromolecule | Name: Envelope glycoprotein gp160 / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Molecular weight | Theoretical: 17.146482 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: AVGIGAVFLG FLGAAGSTMG AASMTLTVQA RNLLSGIVQQ QSNLLRAPEA QQHLLKLTVW GIKQLQARVL AVERYLRDQQ LLGIWGCSG KLICCTNVPW NSSWSNRNLS EIWDNMTWLQ WDKEISNYTQ IIYGLLEESQ NQQEKNEQDL LALD UniProtKB: Envelope glycoprotein gp160 |
-Macromolecule #3: Monoclonal antibody 10A light chain
| Macromolecule | Name: Monoclonal antibody 10A light chain / type: protein_or_peptide / ID: 3 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 23.047424 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: DIVMTQTPAS VEAAVGGTVA IKCQASQSIR SYLAWYQQKP GQPPKLLIYE ASKLASGVPS RFSGSGSGTQ FTLTISGVEC DDAATYYCQ RNYDSYSGAY YPNGFGGGTE VVVKGDPVAP SVLIFPPAAD QVATGTVTIV CVANKYFPDV TVTWEVDGTT Q TTGIENSK ...String: DIVMTQTPAS VEAAVGGTVA IKCQASQSIR SYLAWYQQKP GQPPKLLIYE ASKLASGVPS RFSGSGSGTQ FTLTISGVEC DDAATYYCQ RNYDSYSGAY YPNGFGGGTE VVVKGDPVAP SVLIFPPAAD QVATGTVTIV CVANKYFPDV TVTWEVDGTT Q TTGIENSK TPQNSADCTY NLSSTLTLTS TQYNSHKEYT CKVTQGTTSV VQSFNRGDC UniProtKB: Ig kappa-b4 chain C region |
-Macromolecule #4: Monoclonal antibody 10A heavy chain
| Macromolecule | Name: Monoclonal antibody 10A heavy chain / type: protein_or_peptide / ID: 4 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 22.263018 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QLVESGGGLV QPGASLTLTC TASGFSFSSD YYMCWVRQAP GKGLEWIACI WTANSISYYA RWAKGRFTIS KTSSTTVTLQ MTSLTAADT ATYFCARGGS GDGQSLWGPG TLVTVSSGQP KAPSVFPLAP CCGDTPSSTV TLGCLVKGYL PEPVTVTWNS G TLTNGVRT ...String: QLVESGGGLV QPGASLTLTC TASGFSFSSD YYMCWVRQAP GKGLEWIACI WTANSISYYA RWAKGRFTIS KTSSTTVTLQ MTSLTAADT ATYFCARGGS GDGQSLWGPG TLVTVSSGQP KAPSVFPLAP CCGDTPSSTV TLGCLVKGYL PEPVTVTWNS G TLTNGVRT FPSVRQSSGL YSLSSVVSVT SSSQPVTCNV AHPATNTKVD KTVAPS UniProtKB: IgG heavy chain VDJ region |
-Macromolecule #11: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 11 / Number of copies: 21 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 5.6 mg/mL |
|---|---|
| Buffer | pH: 7.4 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 10 sec. |
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3710 pixel / Digitization - Dimensions - Height: 3838 pixel / Average exposure time: 8.5 sec. / Average electron dose: 52.6 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.5 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
|---|---|
| Output model |  PDB-6did: |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















