+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-7520 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | cryo-EM reconstruction of microtubule-bound 2R-Tau | |||||||||
 Map data Map data | cryo-EM reconstruction of shortened tau construct (WT residues: 197-304,368-400) corresponding to the first two microtubule-binding repeat sequences. | |||||||||
 Sample Sample |
| |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | helical reconstruction / cryo EM / Resolution: 5.6 Å | |||||||||
 Authors Authors | Nogales E / Hejab NMA / Kellogg EH | |||||||||
 Citation Citation |  Journal: Science / Year: 2018 Journal: Science / Year: 2018Title: Near-atomic model of microtubule-tau interactions. Authors: Elizabeth H Kellogg / Nisreen M A Hejab / Simon Poepsel / Kenneth H Downing / Frank DiMaio / Eva Nogales /  Abstract: Tau is a developmentally regulated axonal protein that stabilizes and bundles microtubules (MTs). Its hyperphosphorylation is thought to cause detachment from MTs and subsequent aggregation into ...Tau is a developmentally regulated axonal protein that stabilizes and bundles microtubules (MTs). Its hyperphosphorylation is thought to cause detachment from MTs and subsequent aggregation into fibrils implicated in Alzheimer's disease. It is unclear which tau residues are crucial for tau-MT interactions, where tau binds on MTs, and how it stabilizes them. We used cryo-electron microscopy to visualize different tau constructs on MTs and computational approaches to generate atomic models of tau-tubulin interactions. The conserved tubulin-binding repeats within tau adopt similar extended structures along the crest of the protofilament, stabilizing the interface between tubulin dimers. Our structures explain the effect of phosphorylation on MT affinity and lead to a model of tau repeats binding in tandem along protofilaments, tethering together tubulin dimers and stabilizing polymerization interfaces. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_7520.map.gz emd_7520.map.gz | 481.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-7520-v30.xml emd-7520-v30.xml emd-7520.xml emd-7520.xml | 16.9 KB 16.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_7520.png emd_7520.png | 423.1 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-7520 http://ftp.pdbj.org/pub/emdb/structures/EMD-7520 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7520 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7520 | HTTPS FTP |
-Related structure data
| Related structure data |  7522C  7523C  7769C  7771C  6cvjC  6cvnC C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_7520.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_7520.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | cryo-EM reconstruction of shortened tau construct (WT residues: 197-304,368-400) corresponding to the first two microtubule-binding repeat sequences. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.32 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
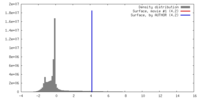
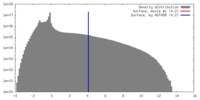
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Ternary complex of alpha-beta tubulin with shortened tau construct
| Entire | Name: Ternary complex of alpha-beta tubulin with shortened tau construct |
|---|---|
| Components |
|
-Supramolecule #1: Ternary complex of alpha-beta tubulin with shortened tau construct
| Supramolecule | Name: Ternary complex of alpha-beta tubulin with shortened tau construct type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: Tau construct contains first two microtubule-targeting repeat motifs and portions of projection-domain and R' sequences |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: 2R-tau
| Macromolecule | Name: 2R-tau / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  |
| Sequence | String: YSSPGSPGTP GSRSRTPSLP TPPTREPKKV AVVRTPPKSP SSAKSRLQTA PVPMPDLKNV KSKIGSTENL KHQPGGGKVQ IINKKLDLSN VQSKCGSKDN IKHVPGGGNK KIETHKLTFR ENAKAKTDHG AEIVYKSPVV S |
-Macromolecule #2: alpha-tubulin
| Macromolecule | Name: alpha-tubulin / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Sequence | String: MRECISIHVG QAGVQIGNAC WELYCLEHGI QPDGQMPSDK TIGGGDDSFN TFFSETGAGK HVPRAVFVDL EPTVIDEVRT GTYRQLFHPE QLITGKEDAA NNYARGHYTI GKEIIDLVLD RIRKLADQCT GLQGFLVFHS FGGGTGSGFT SLLMERLSVD YGKKSKLEFS ...String: MRECISIHVG QAGVQIGNAC WELYCLEHGI QPDGQMPSDK TIGGGDDSFN TFFSETGAGK HVPRAVFVDL EPTVIDEVRT GTYRQLFHPE QLITGKEDAA NNYARGHYTI GKEIIDLVLD RIRKLADQCT GLQGFLVFHS FGGGTGSGFT SLLMERLSVD YGKKSKLEFS IYPAPQVSTA VVEPYNSILT THTTLEHSDC AFMVDNEAIY DICRRNLDIE RPTYTNLNRL ISQIVSSITA SLRFDGALNV DLTEFQTNLV PYPRIHFPLA TYAPVISAEK AYHEQLSVAE ITNACFEPAN QMVKCDPRHG KYMACCLLYR GDVVPKDVNA AIATIKTKRS IQFVDWCPTG FKVGINYQPP TVVPGGDLAK VQRAVCMLSN TTAIAEAWAR LDHKFDLMYA KRAFVHWYVG EGMEEGEFSE AREDMAALEK DYEEVGVDS |
-Macromolecule #3: beta-tubulin
| Macromolecule | Name: beta-tubulin / type: protein_or_peptide / ID: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Sequence | String: MREIVHIQAG QCGNQIGAKF WEVISDEHGI DPTGSYHGDS DLQLERINVY YNEAAGNKYV PRAILVDLEP GTMDSVRSGP FGQIFRPDNF VFGQSGAGNN WAKGHYTEGA ELVDSVLDVV RKESESCDCL QGFQLTHSLG GGTGSGMGTL LISKIREEYP DRIMNTFSVV ...String: MREIVHIQAG QCGNQIGAKF WEVISDEHGI DPTGSYHGDS DLQLERINVY YNEAAGNKYV PRAILVDLEP GTMDSVRSGP FGQIFRPDNF VFGQSGAGNN WAKGHYTEGA ELVDSVLDVV RKESESCDCL QGFQLTHSLG GGTGSGMGTL LISKIREEYP DRIMNTFSVV PSPKVSDTVV EPYNATLSVH QLVENTDETY CIDNEALYDI CFRTLKLTTP TYGDLNHLVS ATMSGVTTCL RFPGQLNADL RKLAVNMVPF PRLHFFMPGF APLTSRGSQQ YRALTVPELT QQMFDAKNMM AACDPRHGRY LTVAAVFRGR MSMKEVDEQM LNVQNKNSSY FVEWIPNNVK TAVCDIPPRG LKMSATFIGN STAIQELFKR ISEQFTAMFR RKAFLHWYTG EGMDEMEFTE AESNMNDLVS EYQQYQDATA D |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Concentration | 0.5 mg/mL | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 6.8 Component:
| ||||||||||||||||||
| Grid | Model: C-flat-1.2/1.3 4C / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR / Details: upspecified | ||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 310.15 K / Instrument: FEI VITROBOT MARK IV Details: blot force 10 pN, 6 second blot time. used C-flat 1.2/1.3 holey grids. First 2 uL of microtubules were adhered to grid for 30 seconds, followed by 2 4 uL washes with 30 second incubation for each wash.. | ||||||||||||||||||
| Details | tubulin concentration is 0.5 mg/mL, tau concentration is 1 mg/mL |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 1 / Number real images: 310 / Average exposure time: 6.0 sec. / Average electron dose: 27.5 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 27500 |
| Sample stage | Specimen holder model: GATAN 626 SINGLE TILT LIQUID NITROGEN CRYO TRANSFER HOLDER Cooling holder cryogen: NITROGEN |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)