[English] 日本語
 Yorodumi
Yorodumi- EMDB-7029: CryoEM map from poorly ordered myosin thick filaments isolated fr... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-7029 | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM map from poorly ordered myosin thick filaments isolated from asynchronous flight muscle of the large waterbug Lethocerus indicus | ||||||||||||||||||
 Map data Map data | Primary map | ||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
| Biological species |  Lethocerus indicus (insect) Lethocerus indicus (insect) | ||||||||||||||||||
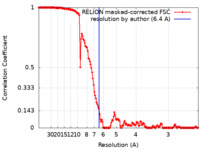
| Method | single particle reconstruction / cryo EM / Resolution: 6.4 Å | ||||||||||||||||||
 Authors Authors | Taylor KA / Taylor D / Hu Z / Edwards RJ | ||||||||||||||||||
| Funding support |  United States, 5 items United States, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: J Struct Biol / Year: 2017 Journal: J Struct Biol / Year: 2017Title: Coupling between myosin head conformation and the thick filament backbone structure. Authors: Zhongjun Hu / Dianne W Taylor / Robert J Edwards / Kenneth A Taylor /   Abstract: The recent high-resolution structure of the thick filament from Lethocerus asynchronous flight muscle shows aspects of thick filament structure never before revealed that may shed some light on how ...The recent high-resolution structure of the thick filament from Lethocerus asynchronous flight muscle shows aspects of thick filament structure never before revealed that may shed some light on how striated muscles function. The phenomenon of stretch activation underlies the function of asynchronous flight muscle. It is most highly developed in flight muscle, but is also observed in other striated muscles such as cardiac muscle. Although stretch activation is likely to be complex, involving more than a single structural aspect of striated muscle, the thick filament itself, would be a prime site for regulatory function because it must bear all of the tension produced by both its associated myosin motors and any externally applied force. Here we show the first structural evidence that the arrangement of myosin heads within the interacting heads motif is coupled to the structure of the thick filament backbone. We find that a change in helical angle of 0.16° disorders the blocked head preferentially within the Lethocerus interacting heads motif. This observation suggests a mechanism for how tension affects the dynamics of the myosin heads leading to a detailed hypothesis for stretch activation and shortening deactivation, in which the blocked head preferentially binds the thin filament followed by the free head when force production occurs. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_7029.map.gz emd_7029.map.gz | 285.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-7029-v30.xml emd-7029-v30.xml emd-7029.xml emd-7029.xml | 15.1 KB 15.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_7029_fsc.xml emd_7029_fsc.xml | 16.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_7029.png emd_7029.png | 165.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-7029 http://ftp.pdbj.org/pub/emdb/structures/EMD-7029 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7029 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7029 | HTTPS FTP |
-Related structure data
| Related structure data |  6so3M M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_7029.map.gz / Format: CCP4 / Size: 307.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_7029.map.gz / Format: CCP4 / Size: 307.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Primary map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.223 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Lethocerus flight muscle myosin filament
| Entire | Name: Lethocerus flight muscle myosin filament |
|---|---|
| Components |
|
-Supramolecule #1: Lethocerus flight muscle myosin filament
| Supramolecule | Name: Lethocerus flight muscle myosin filament / type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  Lethocerus indicus (insect) / Organ: Dorsal longitudinal flight muscle / Tissue: striated muscle / Organelle: myofibril Lethocerus indicus (insect) / Organ: Dorsal longitudinal flight muscle / Tissue: striated muscle / Organelle: myofibril |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 6.8 |
|---|---|
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Support film - Material: CARBON / Support film - topology: HOLEY ARRAY / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Atmosphere: OTHER / Details: Gatan Solarus 950.M plasma cleaner |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 295 K / Instrument: FEI VITROBOT MARK IV |
| Details | All specimen and sample preparation details may be found in Z. Hu, D. W. Taylor, M. K. Reedy, R. J. Edwards, K. A. Taylor, Structure of myosin filaments from relaxed Lethocerus flight muscle by cryo-EM at 6 Angstrom resolution. Sci. Adv. 2, e1600058 (2016). Can also be found in the Specimen section of EMD-3301 |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Details | All data collection details may be found in Z. Hu, D. W. Taylor, M. K. Reedy, R. J. Edwards, K. A. Taylor, Structure of myosin filaments from relaxed Lethocerus flight muscle by cryo-EM at 6 Angstrom resolution. Sci. Adv. 2, e1600058 (2016). Can also be found in the Specimen section of EMD-3301 |
| Image recording | Film or detector model: DIRECT ELECTRON DE-20 (5k x 3k) / Detector mode: INTEGRATING / Digitization - Frames/image: 1-48 / Average exposure time: 1.5 sec. / Average electron dose: 65.0 e/Å2 Details: All specimen and sample preparation details may be found in Z. Hu, D. W. Taylor, M. K. Reedy, R. J. Edwards, K. A. Taylor, Structure of myosin filaments from relaxed Lethocerus flight muscle ...Details: All specimen and sample preparation details may be found in Z. Hu, D. W. Taylor, M. K. Reedy, R. J. Edwards, K. A. Taylor, Structure of myosin filaments from relaxed Lethocerus flight muscle by cryo-EM at 6 Angstrom resolution. Sci. Adv. 2, e1600058 (2016). Can also be found in the Specimen section of EMD-3301 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.5 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: RIGID BODY FIT |
|---|---|
| Output model |  PDB-6so3: |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)