[English] 日本語
 Yorodumi
Yorodumi- EMDB-3301: The Structure of the Relaxed Thick Filaments from Lethocerus Flig... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-3301 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | The Structure of the Relaxed Thick Filaments from Lethocerus Flight Muscle | |||||||||
 Map data Map data | CryoEM IHRSR of relaxed thick filament from Lethocerus indicus (water bug) flight muscle | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | thick filament / insect fight muscle / myosin / paramyosin / myofilin / flightin / relaxed state / muscle contraction | |||||||||
| Function / homology | Myofilin / Myofilin / Myofilin / Flightin / Myofilin protein Function and homology information Function and homology information | |||||||||
| Biological species |  Lethocerus indicus (insect) Lethocerus indicus (insect) | |||||||||
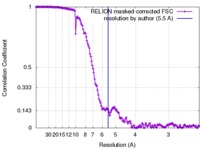
| Method | single particle reconstruction / cryo EM / negative staining / Resolution: 5.5 Å | |||||||||
 Authors Authors | Hu Z / Taylor DW / Reedy MK / Edwards RJ / Taylor KA | |||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2016 Journal: Sci Adv / Year: 2016Title: Structure of myosin filaments from relaxed flight muscle by cryo-EM at 6 Å resolution. Authors: Zhongjun Hu / Dianne W Taylor / Michael K Reedy / Robert J Edwards / Kenneth A Taylor /  Abstract: We describe a cryo-electron microscopy three-dimensional image reconstruction of relaxed myosin II-containing thick filaments from the flight muscle of the giant water bug . The relaxed thick ...We describe a cryo-electron microscopy three-dimensional image reconstruction of relaxed myosin II-containing thick filaments from the flight muscle of the giant water bug . The relaxed thick filament structure is a key element of muscle physiology because it facilitates the reextension process following contraction. Conversely, the myosin heads must disrupt their relaxed arrangement to drive contraction. Previous models predicted that myosin was unique in having an intermolecular head-head interaction, as opposed to the intramolecular head-head interaction observed in all other species. In contrast to the predicted model, we find an intramolecular head-head interaction, which is similar to that of other thick filaments but oriented in a distinctly different way. The arrangement of myosin's long α-helical coiled-coil rod domain has been hypothesized as either curved layers or helical subfilaments. Our reconstruction is the first report having sufficient resolution to track the rod α helices in their native environment at resolutions ~5.5 Å, and it shows that the layer arrangement is correct for . Threading separate paths through the forest of myosin coiled coils are four nonmyosin peptides. We suggest that the unusual position of the heads and the rod arrangement separated by nonmyosin peptides are adaptations for mechanical signal transduction whereby applied tension disrupts the myosin heads as a component of stretch activation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_3301.map.gz emd_3301.map.gz | 283.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-3301-v30.xml emd-3301-v30.xml emd-3301.xml emd-3301.xml | 15.4 KB 15.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_3301_fsc.xml emd_3301_fsc.xml | 15.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_3301.png emd_3301.png | 198.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-3301 http://ftp.pdbj.org/pub/emdb/structures/EMD-3301 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3301 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3301 | HTTPS FTP |
-Related structure data
| Related structure data | |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_3301.map.gz / Format: CCP4 / Size: 300.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_3301.map.gz / Format: CCP4 / Size: 300.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | CryoEM IHRSR of relaxed thick filament from Lethocerus indicus (water bug) flight muscle | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.223 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
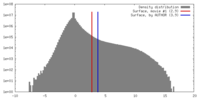
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Thick filament from Lethocerus (waterbug) flight muscle
| Entire | Name: Thick filament from Lethocerus (waterbug) flight muscle |
|---|---|
| Components |
|
-Supramolecule #1000: Thick filament from Lethocerus (waterbug) flight muscle
| Supramolecule | Name: Thick filament from Lethocerus (waterbug) flight muscle type: sample / ID: 1000 Details: The sample is a bipolar helical structure, with helical repeat 145 Angstrom and helical turn 33.98 degree. The sample has C4 symmetry. The map contains 6 unique features: myosin molecule ...Details: The sample is a bipolar helical structure, with helical repeat 145 Angstrom and helical turn 33.98 degree. The sample has C4 symmetry. The map contains 6 unique features: myosin molecule with completely resolved rods, 4 resolved non-myosin densities among the myosin rods and an annular region inside of annulus occupied by myosin rods that most likely contains paramyosin. The 4 non-myosin densities may contain parts of the proteins myofilin and flightin. Oligomeric state: bipolar helical structure / Number unique components: 4 |
|---|
-Macromolecule #1: myosin II
| Macromolecule | Name: myosin II / type: protein_or_peptide / ID: 1 Details: The resolution of LMM and C-terminal of S2 domain is about 5.5 Angstrom; the N-terminal (at first crown) of S2 is about 7 angstrom; free head is about 10 angstrom; block head is about 20 angstrom. Oligomeric state: Helical assembly / Recombinant expression: No / Database: NCBI |
|---|---|
| Source (natural) | Organism:  Lethocerus indicus (insect) / synonym: giant waterbug / Tissue: dorsal longitudinal indirect flight muscle / Cell: myocyte / Organelle: sarcomere / Location in cell: myofibril Lethocerus indicus (insect) / synonym: giant waterbug / Tissue: dorsal longitudinal indirect flight muscle / Cell: myocyte / Organelle: sarcomere / Location in cell: myofibril |
| Molecular weight | Theoretical: 520 KDa |
-Macromolecule #2: paramyosin
| Macromolecule | Name: paramyosin / type: protein_or_peptide / ID: 2 Details: Paramyosin is located in the filament core, and may not have the same symmetry with myosin. The resolution is lower at 10 Angstrom. Oligomeric state: dimer / Recombinant expression: No / Database: NCBI |
|---|---|
| Source (natural) | Organism:  Lethocerus indicus (insect) / synonym: giant waterbug / Tissue: dorsal longitudinal indirect flight muscle / Cell: myocyte / Organelle: sarcomere / Location in cell: myofibril Lethocerus indicus (insect) / synonym: giant waterbug / Tissue: dorsal longitudinal indirect flight muscle / Cell: myocyte / Organelle: sarcomere / Location in cell: myofibril |
| Molecular weight | Theoretical: 100 KDa |
-Macromolecule #3: flightin
| Macromolecule | Name: flightin / type: protein_or_peptide / ID: 3 Details: We can see some non-myosin densities in the reconstruction. Their identification as flightin is uncertain Recombinant expression: No / Database: NCBI |
|---|---|
| Source (natural) | Organism:  Lethocerus indicus (insect) / synonym: giant waterbug / Tissue: dorsal longitudinal indirect flight muscle / Cell: myocyte / Organelle: sarcomere / Location in cell: myofibril Lethocerus indicus (insect) / synonym: giant waterbug / Tissue: dorsal longitudinal indirect flight muscle / Cell: myocyte / Organelle: sarcomere / Location in cell: myofibril |
| Molecular weight | Theoretical: 19 KDa |
| Sequence | UniProtKB: Flightin |
-Macromolecule #4: myofilin
| Macromolecule | Name: myofilin / type: protein_or_peptide / ID: 4 Details: We can see some non-myosin densities in the reconstruction. Their identification as myofilin is uncertain. Recombinant expression: No / Database: NCBI |
|---|---|
| Source (natural) | Organism:  Lethocerus indicus (insect) / synonym: giant waterbug / Tissue: dorsal longitudinal indirect flight muscle / Cell: myocyte / Organelle: sarcomere / Location in cell: myofibril Lethocerus indicus (insect) / synonym: giant waterbug / Tissue: dorsal longitudinal indirect flight muscle / Cell: myocyte / Organelle: sarcomere / Location in cell: myofibril |
| Molecular weight | Theoretical: 30.3 KDa |
| Sequence | UniProtKB: Myofilin protein / InterPro: Myofilin |
-Experimental details
-Structure determination
| Method | negative staining, cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 6.8 Details: 8 mM MOPS, 10 mM Na acetate, 1 mM Mg acetate, 1 mM ATP, and 1 mM EGTA |
|---|---|
| Staining | Type: NEGATIVE Details: All reconstruction data obtained from unstained, frozen hydrated samples. |
| Grid | Details: R2/1 Quantifoil grid |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 100 K / Instrument: FEI VITROBOT MARK IV Details: The Vitrobot environmental chamber was set to 100% relative humidity for freezing and maintained at a temperature of 22 degrees Celsius. The crude filament prep was applied to the grid, ...Details: The Vitrobot environmental chamber was set to 100% relative humidity for freezing and maintained at a temperature of 22 degrees Celsius. The crude filament prep was applied to the grid, incubated for 60 seconds, blotted once and then plunged into liquid ethane to vitrify. Method: EM samples of relaxed filaments were prepared by applying 4 microliters of the above prep to a R2/1 Quantifoil grid at room temperature for 1 minute, washing with several drops of rinse ...Method: EM samples of relaxed filaments were prepared by applying 4 microliters of the above prep to a R2/1 Quantifoil grid at room temperature for 1 minute, washing with several drops of rinse buffer, identical to the relaxing buffer except NaCl was replaced with Na acetate. Following a 2 minute incubation, the grid was washed with several drops of a low salt rinse consisting of 8 mM MOPS, 10 mM Na Acetate, 1 mM Mg Acetate, 1 mM ATP, and 1 mM EGTA, pH 6.8. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Temperature | Min: 90 K |
| Details | 48 frames were recorded over a 1.5 sec exposure time |
| Date | Mar 27, 2015 |
| Image recording | Category: CCD / Film or detector model: OTHER / Number real images: 4000 / Average electron dose: 65 e/Å2 / Details: The total dose is 65 electrons with 48 frames / Bits/pixel: 32 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 29000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)