[English] 日本語
 Yorodumi
Yorodumi- EMDB-61603: Cryo-EM structure of CN-HedgehogCoV (HKU31/Erinaceus amurensis/Ch... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of CN-HedgehogCoV (HKU31/Erinaceus amurensis/China/2014) S-trimer in a locked-2 conformation | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | spike protein / VIRAL PROTEIN | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationvirion component / host cell endoplasmic reticulum-Golgi intermediate compartment membrane / receptor-mediated virion attachment to host cell / endocytosis involved in viral entry into host cell / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / host cell plasma membrane / virion membrane / membrane Similarity search - Function | ||||||||||||
| Biological species |  Erinaceus hedgehog coronavirus HKU31 / Erinaceus hedgehog coronavirus HKU31 /  Enterobacteria phage T4 (virus) Enterobacteria phage T4 (virus) | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.3 Å | ||||||||||||
 Authors Authors | Yuan H / Xiong X | ||||||||||||
| Funding support | 3 items
| ||||||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2025 Journal: Sci Adv / Year: 2025Title: Structures and receptor binding activities of merbecovirus spike proteins reveal key signatures for human DPP4 adaptation. Authors: Hang Yuan / Jingjing Wang / Yong Ma / Zimu Li / Xijie Gao / Gul Habib / Banghui Liu / Jing Chen / Jun He / Peng Zhou / Zheng-Li Shi / Xinwen Chen / Xiaoli Xiong /  Abstract: Merbecoviruses from bats, pangolins, and hedgehogs pose significant zoonotic threats, with a limited understanding of receptor binding by their spike (S) proteins. Here, we report cryo-EM structures ...Merbecoviruses from bats, pangolins, and hedgehogs pose significant zoonotic threats, with a limited understanding of receptor binding by their spike (S) proteins. Here, we report cryo-EM structures of GD-BatCoV (BtCoV-422) and SE-PangolinCoV (MjHKU4r-CoV-1) RBDs in complex with human DPP4 (hDPP4). These structures exhibit a substantial offset in their hDPP4 interaction interfaces, revealing a conserved hydrophobic cluster as a convergent signature of DPP4 binding within the MERS-HKU4 clade of merbecoviruses. Structure-guided mutagenesis demonstrates that favorable interactions are distributed across multiple receptor binding motif (RBM) regions, working synergistically to confer high-affinity hDPP4 binding. Swapping of the merbecovirus RBM regions indicate limited plasticity and interchangeability among these regions. In addition, we report cryo-EM structures of six merbecovirus S-trimers. Structure-based phylogenetics suggests that hDPP4-binding merbecoviruses undergo convergent evolution, while ACE2-binding merbecoviruses exhibit diversification in their binding mechanisms. These findings offer critical insights into merbecovirus receptor utilization, providing a structural understanding for future surveillance. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_61603.map.gz emd_61603.map.gz | 49.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-61603-v30.xml emd-61603-v30.xml emd-61603.xml emd-61603.xml | 19.9 KB 19.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_61603.png emd_61603.png | 12.4 KB | ||
| Filedesc metadata |  emd-61603.cif.gz emd-61603.cif.gz | 7 KB | ||
| Others |  emd_61603_half_map_1.map.gz emd_61603_half_map_1.map.gz emd_61603_half_map_2.map.gz emd_61603_half_map_2.map.gz | 40.9 MB 40.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-61603 http://ftp.pdbj.org/pub/emdb/structures/EMD-61603 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-61603 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-61603 | HTTPS FTP |
-Related structure data
| Related structure data |  9jmiMC  9jmfC  9jmgC  9jmhC  9jmjC  9jmmC  9jmnC  9jmoC  9jmpC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_61603.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_61603.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.32 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_61603_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_61603_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : CN-HedgehogCoV (HKU31/Erinaceus amurensis/China/2014) Spike protein
| Entire | Name: CN-HedgehogCoV (HKU31/Erinaceus amurensis/China/2014) Spike protein |
|---|---|
| Components |
|
-Supramolecule #1: CN-HedgehogCoV (HKU31/Erinaceus amurensis/China/2014) Spike protein
| Supramolecule | Name: CN-HedgehogCoV (HKU31/Erinaceus amurensis/China/2014) Spike protein type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Erinaceus hedgehog coronavirus HKU31 Erinaceus hedgehog coronavirus HKU31 |
-Macromolecule #1: Spike glycoprotein,Fibritin
| Macromolecule | Name: Spike glycoprotein,Fibritin / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Enterobacteria phage T4 (virus) Enterobacteria phage T4 (virus) |
| Molecular weight | Theoretical: 149.577219 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MIRSVFLLMC LLTLIKSGDA NCIVVDMQPS YFQRDWPRPI NMSAADGIIY PVGRTYANIT IMLSGLFPHQ ADKGKQYIYS MYHMKFNQG VNDPFISNYS YYTEPFDNGF VVRIGANSNI TGSSVVGSAN ATVKKIYPAL MLGSVVGKFP SNKTGRYFNH T LVILPEKC ...String: MIRSVFLLMC LLTLIKSGDA NCIVVDMQPS YFQRDWPRPI NMSAADGIIY PVGRTYANIT IMLSGLFPHQ ADKGKQYIYS MYHMKFNQG VNDPFISNYS YYTEPFDNGF VVRIGANSNI TGSSVVGSAN ATVKKIYPAL MLGSVVGKFP SNKTGRYFNH T LVILPEKC GTQITALYCV LQPKNQTNCT GHTTYTSYVL VENKTCSTDA DGNLKQSLQT WFDLKDCLFE KKYNVSADER EE WFGITQD QQGVHLYTSR KNGFKSNNMF LFATLPIYER LRYYTVMPRS IKTTANNDHM AWSAFYIYQL HKLNYLVEFD VEG YIVRAS DCGANDYTQL QCSYGQFDMN SGVYSASYFN AQPRGYYYVA NELEECALDV LFKNIAPQIA NYSRRVFTNC NYNL TKLLS LVEVDEFVCD KTTPESLATG CYSSLVVDWF ALPLSMKSTL AIGSAEAISM FNYNQDYSNP TCRIHATINS NVSSS LNFT ANNNYAYISR CQGTDGKPIL LQKGQLPNIA CRSGVLGLSN DVDYFGYNFN GRIFYIGRKS YTPRTSEGSI QMVYVI TAN YAEGPNNVCP LKDTTTATDN LDSLLGQCID YDINGVVGQG VFQKCNSTGI PSQVFVYDSF HNIIGYHSKN GTYYCMA PC VSVPVSVIYD KSSNSHATLY GSVACDHIKM IPSVLSRETA TKLRASDNGL LQTAVGCVIG FHNTSDTVED CNLPLGQS L CAKPPSLTRS ANNTFGLAVM KYENPLKVEV LNSSEFEISV PTNFSFRVTE EFIETSIQKV TVDCKQYVCN GFERCEQLL EQYGQFCAKI NQALHGVNLR QDDATKSLFE SIRVPQSAPL MASLSGDYNL SLFETPSINT GGSGNYRSAL EDLLFDKVTL SVPGYMKGY DECMKKGPPS ARDLICAQYV AGYKVLPPLY DSNMEAMYTA SLTGSIAGAL WTGGLSSAAS LPFAQSIYYR M NGIGITQN VLMENQKLIA NKFNQALGAM QTGFTTTNQA FQKVQDAVNA NAQALSKLAS ELSNTFGAIS ASIGDILKRL DT LEQEVQI DRLINGRLTS LNAFVSQQLV RSEAAARSSQ LVKDKVNECV KSQSSRNGFC GQGTHIVSFA VNAPNGFYFV HVG YHPTDY VNQTSAYGLC DSSNTKCIAA KNGYFIKNGS DDWMFTGSSY YQPEPITNFN SRSVEPEVTF QNLTNNLPPP LSNS TDVDF KDELEEFFKN ITSEVPNFGS ITQINSTVLD LSEEMKTLQS VVEALNQSYI ELKELGNYTY YNKWPGSGYI PEAPR DGQA YVRKDGEWVL LSTFLLEVLF QGPGHHHHHH HHSAWSHPQF EKGGGSGGGG SGGSAWSHPQ FEKSA UniProtKB: Spike glycoprotein, Fibritin |
-Macromolecule #4: FOLIC ACID
| Macromolecule | Name: FOLIC ACID / type: ligand / ID: 4 / Number of copies: 3 / Formula: FOL |
|---|---|
| Molecular weight | Theoretical: 441.397 Da |
| Chemical component information | 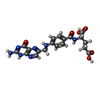 ChemComp-FA: |
-Macromolecule #5: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 5 / Number of copies: 39 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





































