[English] 日本語
 Yorodumi
Yorodumi- EMDB-61601: Cryo-EM structure of EU-HedgehogCoV (Erinaceus/VMC/DEU/2012) S-tr... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of EU-HedgehogCoV (Erinaceus/VMC/DEU/2012) S-trimer in a locked-2 conformation | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | spike protein / VIRAL PROTEIN | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationvirion component / host cell endoplasmic reticulum-Golgi intermediate compartment membrane / receptor-mediated virion attachment to host cell / endocytosis involved in viral entry into host cell / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / host cell plasma membrane / virion membrane / membrane Similarity search - Function | ||||||||||||
| Biological species |  Betacoronavirus Erinaceus/VMC/DEU/2012 / Betacoronavirus Erinaceus/VMC/DEU/2012 /  Enterobacteria phage T4 (Bacteriophage T4) (virus) Enterobacteria phage T4 (Bacteriophage T4) (virus) | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.0 Å | ||||||||||||
 Authors Authors | Yuan H / Xiong X | ||||||||||||
| Funding support | 3 items
| ||||||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2025 Journal: Sci Adv / Year: 2025Title: Structures and receptor binding activities of merbecovirus spike proteins reveal key signatures for human DPP4 adaptation. Authors: Hang Yuan / Jingjing Wang / Yong Ma / Zimu Li / Xijie Gao / Gul Habib / Banghui Liu / Jing Chen / Jun He / Peng Zhou / Zheng-Li Shi / Xinwen Chen / Xiaoli Xiong /  Abstract: Merbecoviruses from bats, pangolins, and hedgehogs pose significant zoonotic threats, with a limited understanding of receptor binding by their spike (S) proteins. Here, we report cryo-EM structures ...Merbecoviruses from bats, pangolins, and hedgehogs pose significant zoonotic threats, with a limited understanding of receptor binding by their spike (S) proteins. Here, we report cryo-EM structures of GD-BatCoV (BtCoV-422) and SE-PangolinCoV (MjHKU4r-CoV-1) RBDs in complex with human DPP4 (hDPP4). These structures exhibit a substantial offset in their hDPP4 interaction interfaces, revealing a conserved hydrophobic cluster as a convergent signature of DPP4 binding within the MERS-HKU4 clade of merbecoviruses. Structure-guided mutagenesis demonstrates that favorable interactions are distributed across multiple receptor binding motif (RBM) regions, working synergistically to confer high-affinity hDPP4 binding. Swapping of the merbecovirus RBM regions indicate limited plasticity and interchangeability among these regions. In addition, we report cryo-EM structures of six merbecovirus S-trimers. Structure-based phylogenetics suggests that hDPP4-binding merbecoviruses undergo convergent evolution, while ACE2-binding merbecoviruses exhibit diversification in their binding mechanisms. These findings offer critical insights into merbecovirus receptor utilization, providing a structural understanding for future surveillance. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_61601.map.gz emd_61601.map.gz | 49.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-61601-v30.xml emd-61601-v30.xml emd-61601.xml emd-61601.xml | 20.5 KB 20.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_61601.png emd_61601.png | 13.9 KB | ||
| Filedesc metadata |  emd-61601.cif.gz emd-61601.cif.gz | 7.1 KB | ||
| Others |  emd_61601_half_map_1.map.gz emd_61601_half_map_1.map.gz emd_61601_half_map_2.map.gz emd_61601_half_map_2.map.gz | 40.9 MB 40.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-61601 http://ftp.pdbj.org/pub/emdb/structures/EMD-61601 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-61601 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-61601 | HTTPS FTP |
-Related structure data
| Related structure data |  9jmgMC  9jmfC  9jmhC  9jmiC  9jmjC  9jmmC  9jmnC  9jmoC  9jmpC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_61601.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_61601.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.32 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_61601_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_61601_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : EU-HedgehogCoV (Erinaceus/VMC/DEU/2012/) Spike protein
| Entire | Name: EU-HedgehogCoV (Erinaceus/VMC/DEU/2012/) Spike protein |
|---|---|
| Components |
|
-Supramolecule #1: EU-HedgehogCoV (Erinaceus/VMC/DEU/2012/) Spike protein
| Supramolecule | Name: EU-HedgehogCoV (Erinaceus/VMC/DEU/2012/) Spike protein type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Betacoronavirus Erinaceus/VMC/DEU/2012 Betacoronavirus Erinaceus/VMC/DEU/2012 |
-Macromolecule #1: Spike glycoprotein,Fibritin
| Macromolecule | Name: Spike glycoprotein,Fibritin / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Enterobacteria phage T4 (Bacteriophage T4) (virus) Enterobacteria phage T4 (Bacteriophage T4) (virus) |
| Molecular weight | Theoretical: 149.867672 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MIRSACLLMC LLMFIKATPS EGTCISVDMQ PSYFIKNWSM PINMSKADGV LYPVGRTYSN ITITLEGLFP HQADRGDMYV YSQSHKGRP FISNYSLVTN DFGNGIVIRI GSAANKTGSP IISNIASNFQ IMKKLYPALL FGSAVGKFPA NNKTGAYFNH S LVILPEKC ...String: MIRSACLLMC LLMFIKATPS EGTCISVDMQ PSYFIKNWSM PINMSKADGV LYPVGRTYSN ITITLEGLFP HQADRGDMYV YSQSHKGRP FISNYSLVTN DFGNGIVIRI GSAANKTGSP IISNIASNFQ IMKKLYPALL FGSAVGKFPA NNKTGAYFNH S LVILPEKC GTVITALYCV LEPKNDTNCT GSSGYVSYVL FENFSCDATT DGNLKNSLQQ WFNIKDCLFE KNYNVTDDER EE WFGIIQD QQGVHLYTSR KNGYSNNMFL FATLPIYDQI LYYTVMPRSI NASNYASYHA FSAFYIYKLH KINYMVDFDV NGY ITRAVD CGYNDYTQLQ CSYGQFDMDS GVYSASYFNA RSRGYYYEAA ELTECDLDVL FKNDAPIIAN YSRRVFTNCN YNLT KLLSL VQVDEFVCHK TTPEALATGC YSSLTVDWFA LPFSMKSTLA IGSAEAISMF NYNQDYSNPT CRIHAAVTAN VSTAL NFTA NANYAYISRC QGVDGKPILL QPGQMTNIAC RSGVLARPSD ADYFGYSFQG RNYYLGRKSY KPKTDEGDVQ MVYVIT PKY DKGPDTVCPL KDMGAASSSL DGLLGQCIDY DIHGVVGRGV FQVCNTTGIA SQIFVYDGFG NIIGFHSKNG TYYCMAP CV SVPVSVIYDK SSDVHATLYS SVECNHIKSV ATVFSRQTES KLRSSDNGLL QTAVGCVIGF RNTSDTVEDC TLPLGQSL C AKPPSFSSRS TNVNNTFALV HMLFQNPLKV SVLNSTEFQV SIPQNFSFGI TEEFIETSIQ KVTVDCKQYV CNGFERCEQ LLVQYGQFCS KINQALHGVN LRQDDATKSL FEDIKVPQSV PLMTSLTGDY NLSLFEAPNI GSGNNNYRSA LEDLLFDKVT LSDPGYMKG YDECMKKGPP SARDLICAQY VSGYKVLPPL YDANMEAMYT ASLTGSIAGS FWTGGLSSAA ALPFAQSMFY R MNGIGITQ NVLMKNQREI ANKFNQALGA MQTGFTATNQ AFQKVQDVVN ANAQALSKLA SELANTFGAI SASIGDILKR LD VLEQEVQ IDRLINGRLT SLNAFVSQQL VRSEAAARSS QLAKEKINEC VKAQSTRSGF CGQGTHIVSF VINAPNGFYF IHV GYHPQD YVNQTAAYGL CDSSSKKCIA AKNGYFVKND SDSGSWSYTG SSFYQPEPIT TFNSRFVEPE VTFQNLSNNL PPPL LSNNT DITFEDELEE FYKNITSEIP NFGSLSQINT TMLNLSQEMS TLQQVVKDLN NSYIELKELG NHTYYQKWPG SGYIP EAPR DGQAYVRKDG EWVLLSTFLL EVLFQGPGHH HHHHHHSAWS HPQFEKGGGS GGGGSGGSAW SHPQFEKSA UniProtKB: Spike glycoprotein, Fibritin |
-Macromolecule #4: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 4 / Number of copies: 37 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Macromolecule #5: FOLIC ACID
| Macromolecule | Name: FOLIC ACID / type: ligand / ID: 5 / Number of copies: 3 / Formula: FOL |
|---|---|
| Molecular weight | Theoretical: 441.397 Da |
| Chemical component information | 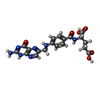 ChemComp-FA: |
-Macromolecule #6: LINOLEIC ACID
| Macromolecule | Name: LINOLEIC ACID / type: ligand / ID: 6 / Number of copies: 3 / Formula: EIC |
|---|---|
| Molecular weight | Theoretical: 280.445 Da |
| Chemical component information |  ChemComp-EIC: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 6 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





































