[English] 日本語
 Yorodumi
Yorodumi- EMDB-4711: Structure of LSD2/NPAC-linker/nucleosome core particle complex: C... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-4711 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of LSD2/NPAC-linker/nucleosome core particle complex: Class 4 | |||||||||
 Map data Map data | LSD/NPAC(214-225)/nucleosome: class 4 | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 6.23 Å | |||||||||
 Authors Authors | Marabelli C / Pilotto S / Chittori S / Subramaniam S / Mattevi A | |||||||||
 Citation Citation |  Journal: Cell Rep / Year: 2019 Journal: Cell Rep / Year: 2019Title: A Tail-Based Mechanism Drives Nucleosome Demethylation by the LSD2/NPAC Multimeric Complex. Authors: Chiara Marabelli / Biagina Marrocco / Simona Pilotto / Sagar Chittori / Sarah Picaud / Sara Marchese / Giuseppe Ciossani / Federico Forneris / Panagis Filippakopoulos / Guy Schoehn / Daniela ...Authors: Chiara Marabelli / Biagina Marrocco / Simona Pilotto / Sagar Chittori / Sarah Picaud / Sara Marchese / Giuseppe Ciossani / Federico Forneris / Panagis Filippakopoulos / Guy Schoehn / Daniela Rhodes / Sriram Subramaniam / Andrea Mattevi /       Abstract: LSD1 and LSD2 are homologous histone demethylases with opposite biological outcomes related to chromatin silencing and transcription elongation, respectively. Unlike LSD1, LSD2 nucleosome-demethylase ...LSD1 and LSD2 are homologous histone demethylases with opposite biological outcomes related to chromatin silencing and transcription elongation, respectively. Unlike LSD1, LSD2 nucleosome-demethylase activity relies on a specific linker peptide from the multidomain protein NPAC. We used single-particle cryoelectron microscopy (cryo-EM), in combination with kinetic and mutational analysis, to analyze the mechanisms underlying the function of the human LSD2/NPAC-linker/nucleosome complex. Weak interactions between LSD2 and DNA enable multiple binding modes for the association of the demethylase to the nucleosome. The demethylase thereby captures mono- and dimethyl Lys4 of the H3 tail to afford histone demethylation. Our studies also establish that the dehydrogenase domain of NPAC serves as a catalytically inert oligomerization module. While LSD1/CoREST forms a nucleosome docking platform at silenced gene promoters, LSD2/NPAC is a multifunctional enzyme complex with flexible linkers, tailored for rapid chromatin modification, in conjunction with the advance of the RNA polymerase on actively transcribed genes. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_4711.map.gz emd_4711.map.gz | 61.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-4711-v30.xml emd-4711-v30.xml emd-4711.xml emd-4711.xml | 15.8 KB 15.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_4711.png emd_4711.png | 119.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-4711 http://ftp.pdbj.org/pub/emdb/structures/EMD-4711 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4711 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4711 | HTTPS FTP |
-Related structure data
| Related structure data |  4704C  4705C  4710C  4712C  6r1tC  6r1uC  6r25C C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_4711.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_4711.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | LSD/NPAC(214-225)/nucleosome: class 4 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
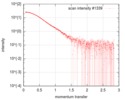
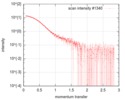
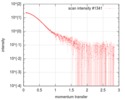
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.06 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : LSD2/NPAC(214-225)/nucleosome
| Entire | Name: LSD2/NPAC(214-225)/nucleosome |
|---|---|
| Components |
|
-Supramolecule #1: LSD2/NPAC(214-225)/nucleosome
| Supramolecule | Name: LSD2/NPAC(214-225)/nucleosome / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: Xenopus laevis histones recombinantly expressed. Alkylated K4C-C110A H3. 601 Widom DNA sequence. Human LSD2 Human NPAC |
|---|---|
| Source (natural) | Organism: |
| Molecular weight | Experimental: 290 KDa |
-Macromolecule #1: LSD2
| Macromolecule | Name: LSD2 / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Sequence | String: PLGSRKCEKA GCTATCPVCF ASASERCAKN GYTSRWYHLS CGEHFCNECF DHYYRSHKDG YDKYTTWKKI WTSNGKTEPS PKAFMADQQ LPYWVQCTKP ECRKWRQLTK EIQLTPQIAK TYRCGMKPNT AIKPETSDHC SLPEDLRVLE VSNHWWYSML I LPPLLKDS ...String: PLGSRKCEKA GCTATCPVCF ASASERCAKN GYTSRWYHLS CGEHFCNECF DHYYRSHKDG YDKYTTWKKI WTSNGKTEPS PKAFMADQQ LPYWVQCTKP ECRKWRQLTK EIQLTPQIAK TYRCGMKPNT AIKPETSDHC SLPEDLRVLE VSNHWWYSML I LPPLLKDS VAAPLLSAYY PDCVGMSPSC TSTNRAAATG NASPGKLEHS KAALSVHVPG MNRYFQPFYQ PNECGKALCV RP DVMELDE LYEFPEYSRD PTMYLALRNL ILALWYTNCK EALTPQKCIP HIIVRGLVRI RCVQEVERIL YFMTRKGLIN TGV LSVGAD QYLLPKDYHN KSVIIIGAGP AGLAAARQLH NFGIKVTVLE AKDRIGGRVW DDKSFKGVTV GRGAQIVNGC INNP VALMC EQLGISMHKF GERCDLIQEG GRITDPTIDK RMDFHFNALL DVVSEWRKDK TQLQDVPLGE KIEEIYKAFI KESGI QFSE LEGQVLQFHL SNLEYACGSN LHQVSARSWD HNEFFAQFAG DHTLLTPGYS VIIEKLAEGL DIQLKSPVQC IDYSGD EVQ VTTTDGTGYS AQKVLVTVPL ALLQKGAIQF NPPLSEKKMK AINSLGAGII EKIALQFPYR FWDSKVQGAD FFGHVPP SA SKRGLFAVFY DMDPQKKHSV LMSVIAGEAV ASVRTLDDKQ VLQQCMATLR ELFKEQEVPD PTKYFVTRWS TDPWIQMA Y SFVKTGGSGE AYDIIAEDIQ GTVFFAGEAT NRHFPQTVTG AYLSGVREAS KIAAF |
-Macromolecule #2: NPAC
| Macromolecule | Name: NPAC / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: DPHFHHFLLS QT |
-Macromolecule #3: H3
| Macromolecule | Name: H3 / type: protein_or_peptide / ID: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  |
| Sequence | String: ARTKQTARKS TGGKAPRKQL ATKAARKSAP ATGGVKKPHR YRPGTVALRE IRRYQKSTEL LIRKLPFQRL VREIAQDFKT DLRFQSSAV MALQEASEAY LVALFEDTNL CAIHAKRVTI MPKDIQLARR IRGERA |
-Macromolecule #4: H4
| Macromolecule | Name: H4 / type: protein_or_peptide / ID: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  |
| Sequence | String: MSGRGKGGKG LGKGGAKRHR KVLRDNIQGI TKPAIRRLAR RGGVKRISGL IYEETRGVLK VFLENVIRD AVTYTEHAKR KTVTAMDVVY ALKRQGRTLY GFGG |
-Macromolecule #5: H2A
| Macromolecule | Name: H2A / type: protein_or_peptide / ID: 5 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  |
| Sequence | String: MSGRGKQGGK TRAKAKTRSS RAGLQFPVGR VHRLLRKGNY AERVGAGAPV YLAAVLEYLT AEILELAGN AARDNKKTRI IPRHLQLAVR NDEELNKLLG GVTIAQGGVL PNIQSVLLPK K TESAKSAK SK |
-Macromolecule #6: H2B
| Macromolecule | Name: H2B / type: protein_or_peptide / ID: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  |
| Sequence | String: AKSAPAPKKG SKKAVTKTQK KDGKKRRKTR KESYAIYVYK VLKQVHPDTG ISSKAMSIMN SFVNDVFERI AGEASRLAHY NKRSTITSR EIQTAVRLLL PGELAKHAVS EGTKAVTKYT SAK |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.87 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| |||||||||
| Vitrification | Cryogen name: ETHANE | |||||||||
| Details | LSD2/NPAC(214-225)/nucleosome was monodisperse (gel filtration peak isolation and concentration in buffer described. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 1 / Number real images: 2078 / Average exposure time: 8.0 sec. / Average electron dose: 1.25 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 0.00305 µm / Nominal defocus min: 0.0007 µm / Nominal magnification: 130000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: |
|---|---|
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
-Atomic model buiding 2
| Initial model | PDB ID: |
|---|---|
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)























