[English] 日本語
 Yorodumi
Yorodumi- EMDB-4712: Structure of LSD2/NPAC-linker/nucleosome core particle complex: C... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-4712 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of LSD2/NPAC-linker/nucleosome core particle complex: Class 5 | |||||||||
 Map data Map data | Unfolded nucleosome | |||||||||
 Sample Sample |
| |||||||||
| Biological species | ||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 7.95 Å | |||||||||
 Authors Authors | Marabelli C / Pilotto S / Chittori S / Subramaniam S / Mattevi A | |||||||||
 Citation Citation |  Journal: Cell Rep / Year: 2019 Journal: Cell Rep / Year: 2019Title: A Tail-Based Mechanism Drives Nucleosome Demethylation by the LSD2/NPAC Multimeric Complex. Authors: Chiara Marabelli / Biagina Marrocco / Simona Pilotto / Sagar Chittori / Sarah Picaud / Sara Marchese / Giuseppe Ciossani / Federico Forneris / Panagis Filippakopoulos / Guy Schoehn / Daniela ...Authors: Chiara Marabelli / Biagina Marrocco / Simona Pilotto / Sagar Chittori / Sarah Picaud / Sara Marchese / Giuseppe Ciossani / Federico Forneris / Panagis Filippakopoulos / Guy Schoehn / Daniela Rhodes / Sriram Subramaniam / Andrea Mattevi /       Abstract: LSD1 and LSD2 are homologous histone demethylases with opposite biological outcomes related to chromatin silencing and transcription elongation, respectively. Unlike LSD1, LSD2 nucleosome-demethylase ...LSD1 and LSD2 are homologous histone demethylases with opposite biological outcomes related to chromatin silencing and transcription elongation, respectively. Unlike LSD1, LSD2 nucleosome-demethylase activity relies on a specific linker peptide from the multidomain protein NPAC. We used single-particle cryoelectron microscopy (cryo-EM), in combination with kinetic and mutational analysis, to analyze the mechanisms underlying the function of the human LSD2/NPAC-linker/nucleosome complex. Weak interactions between LSD2 and DNA enable multiple binding modes for the association of the demethylase to the nucleosome. The demethylase thereby captures mono- and dimethyl Lys4 of the H3 tail to afford histone demethylation. Our studies also establish that the dehydrogenase domain of NPAC serves as a catalytically inert oligomerization module. While LSD1/CoREST forms a nucleosome docking platform at silenced gene promoters, LSD2/NPAC is a multifunctional enzyme complex with flexible linkers, tailored for rapid chromatin modification, in conjunction with the advance of the RNA polymerase on actively transcribed genes. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_4712.map.gz emd_4712.map.gz | 62.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-4712-v30.xml emd-4712-v30.xml emd-4712.xml emd-4712.xml | 16.8 KB 16.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_4712.png emd_4712.png | 92.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-4712 http://ftp.pdbj.org/pub/emdb/structures/EMD-4712 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4712 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4712 | HTTPS FTP |
-Related structure data
| Related structure data |  4704C  4705C  4710C  4711C  6r1tC  6r1uC  6r25C C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_4712.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_4712.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unfolded nucleosome | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.06 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
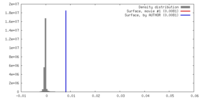
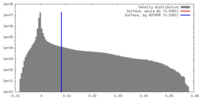
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : partly unfolded nucleosome
| Entire | Name: partly unfolded nucleosome |
|---|---|
| Components |
|
-Supramolecule #1: partly unfolded nucleosome
| Supramolecule | Name: partly unfolded nucleosome / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#8 Details: Xenopus laevis histones recombinantly expressed. Alkylated K4C-C110A H3. 601 Widom DNA sequence. Human LSD2 Human NPAC |
|---|---|
| Source (natural) | Organism: |
| Molecular weight | Theoretical: 178 KDa |
-Macromolecule #1: H3
| Macromolecule | Name: H3 / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Sequence | String: ARTKQTARKS TGGKAPRKQL ATKAARKSAP ATGGVKKPHR YRPGTVALRE IRRYQKSTEL LIRKLPFQRL VREIAQDFKT DLRFQSSAV MALQEASEAY LVALFEDTNL CAIHAKRVTI MPKDIQLARR IRGERA |
-Macromolecule #2: H4
| Macromolecule | Name: H4 / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: |
| Recombinant expression | Organism:  |
| Sequence | String: SGRGKGGKGL GKGGAKRHRK VLRDNIQGIT KPAIRRLARR GGVKRISGLI YEETRGVLKV FLENVIRDAV TYTEHAKRKT VTAMDVVYA LKRQGRTLYG FGG |
-Macromolecule #3: H2A
| Macromolecule | Name: H2A / type: protein_or_peptide / ID: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: |
| Recombinant expression | Organism:  |
| Sequence | String: SGRGKQGGKT RAKAKTRSSR AGLQFPVGRV HRLLRKGNYA ERVGAGAPVY LAAVLEYLTA EILELAGNAA RDNKKTRIIP RHLQLAVRN DEELNKLLGR VTIAQGGVLP NIQSVLLPKK TESSKSAKSK |
-Macromolecule #4: H2B
| Macromolecule | Name: H2B / type: protein_or_peptide / ID: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: |
| Recombinant expression | Organism:  |
| Sequence | String: MPDPAKSAPA AKKGSKKAVT KTQKKDGKKR RKSRKESYAI YVYKVLKQVH PDTGISSKAM SIMNSFVND VFERIAGEAS RLAHYNKRST ITSREIQTAV RLLLPGELAK HAVSEGTKAV T KYTSAK |
-Macromolecule #7: H3
| Macromolecule | Name: H3 / type: protein_or_peptide / ID: 7 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: |
| Recombinant expression | Organism:  |
| Sequence | String: ARTKQTARKS TGGKAPRKQL ATKAARKSAP ATGGVKKPHR YRPGTVALRE IRRYQKSTEL LIRKLPFQRL VREIAQDFKT DLRFQSSAV MALQEASEAY LVALFEDTNL CAIHAKRVTI MPKDIQLARR IRGERA |
-Macromolecule #8: H4
| Macromolecule | Name: H4 / type: protein_or_peptide / ID: 8 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: |
| Recombinant expression | Organism:  |
| Sequence | String: SGRGKGGKGL GKGGAKRHRK VLRDNIQGIT KPAIRRLARR GGVKRISGLI YEETRGVLKV FLENVIRDAV TYTEHAKRKT VTAMDVVYA LKRQGRTLYG FGG |
-Macromolecule #5: DNA (147mer)
| Macromolecule | Name: DNA (147mer) / type: dna / ID: 5 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Sequence | String: ATCGGATGTA TATATCTGAC ACGTGCCTGG AGACTAGGGA GTAATCCCCT TGGCGGTTAA AACGCGGGGG ACAGCGCGTA CGTGCGTTT AAGCGGTGCT AGAGCTGTCT ACGACCAATT GAGCGGCCTC GGCACCGGGA TTCTCGAT |
-Macromolecule #6: DNA (147mer)
| Macromolecule | Name: DNA (147mer) / type: dna / ID: 6 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Sequence | String: ATCGAGAATC CCGGTGCCGA GGCCGCTCAA TTGGTCGTAG ACAGCTCTAG CACCGCTTAA ACGCACGTAC GCGCTGTCCC CCGCGTTTT AACCGCCAAG GGGATTACTC CCTAGTCTCC AGGCACGTGT CAGATATATA CATCCGAT |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.87 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| |||||||||
| Vitrification | Cryogen name: ETHANE | |||||||||
| Details | LSD2/NPAC(214-225)/nucleosome was monodisperse (gel filtration peak isolation and concentration in buffer described. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 1 / Number real images: 2078 / Average exposure time: 8.0 sec. / Average electron dose: 1.25 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 0.00305 µm / Nominal defocus min: 0.0007 µm / Nominal magnification: 130000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: |
|---|---|
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
-Atomic model buiding 2
| Initial model | PDB ID: |
|---|---|
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)