+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-4257 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Additional yeast OST cryoEM maps from focused 3D refinements | |||||||||
 Map data Map data | Luminal part of OST, Post-processed, masked map, "Map 2" | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
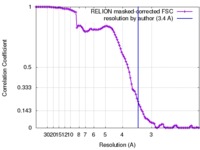
| Method | single particle reconstruction / cryo EM / Resolution: 3.4 Å | |||||||||
 Authors Authors | Wild R / Kowal J / Eyring J / Ngwa EM / Aebi M / Locher KP | |||||||||
 Citation Citation |  Journal: Science / Year: 2018 Journal: Science / Year: 2018Title: Structure of the yeast oligosaccharyltransferase complex gives insight into eukaryotic N-glycosylation. Authors: Rebekka Wild / Julia Kowal / Jillianne Eyring / Elsy M Ngwa / Markus Aebi / Kaspar P Locher /  Abstract: Oligosaccharyltransferase (OST) is an essential membrane protein complex in the endoplasmic reticulum, where it transfers an oligosaccharide from a dolichol-pyrophosphate-activated donor to ...Oligosaccharyltransferase (OST) is an essential membrane protein complex in the endoplasmic reticulum, where it transfers an oligosaccharide from a dolichol-pyrophosphate-activated donor to glycosylation sites of secretory proteins. Here we describe the atomic structure of yeast OST determined by cryo-electron microscopy, revealing a conserved subunit arrangement. The active site of the catalytic STT3 subunit points away from the center of the complex, allowing unhindered access to substrates. The dolichol-pyrophosphate moiety binds to a lipid-exposed groove of STT3, whereas two noncatalytic subunits and an ordered N-glycan form a membrane-proximal pocket for the oligosaccharide. The acceptor polypeptide site faces an oxidoreductase domain in stand-alone OST complexes or is immediately adjacent to the translocon, suggesting how eukaryotic OSTs efficiently glycosylate a large number of polypeptides before their folding. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_4257.map.gz emd_4257.map.gz | 6.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-4257-v30.xml emd-4257-v30.xml emd-4257.xml emd-4257.xml | 14.5 KB 14.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_4257_fsc.xml emd_4257_fsc.xml | 10.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_4257.png emd_4257.png | 206.9 KB | ||
| Others |  emd_4257_additional_1.map.gz emd_4257_additional_1.map.gz emd_4257_additional_2.map.gz emd_4257_additional_2.map.gz emd_4257_additional_3.map.gz emd_4257_additional_3.map.gz | 6 MB 5 MB 4.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-4257 http://ftp.pdbj.org/pub/emdb/structures/EMD-4257 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4257 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4257 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_4257.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_4257.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Luminal part of OST, Post-processed, masked map, "Map 2" | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.065 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: Transmembrane region of OST, Post-processed, masked map, "Map 3"
| File | emd_4257_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Transmembrane region of OST, Post-processed, masked map, "Map 3" | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: OST1 region of OST, Post-processed, masked map, "Map 5"
| File | emd_4257_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | OST1 region of OST, Post-processed, masked map, "Map 5" | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: SWP1 region of OST, Post-processed, masked map, "Map 4"
| File | emd_4257_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | SWP1 region of OST, Post-processed, masked map, "Map 4" | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : yeast OST
| Entire | Name: yeast OST |
|---|---|
| Components |
|
-Supramolecule #1: yeast OST
| Supramolecule | Name: yeast OST / type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE-PROPANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 2.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)