[English] 日本語
 Yorodumi
Yorodumi- EMDB-4161: Cryo-EM structure of the yeast oligosaccharyltransferase (OST) complex -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-4161 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the yeast oligosaccharyltransferase (OST) complex | |||||||||
 Map data Map data | None | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | OST complex / oligosaccharyltransferase / N-linked glycosylation / yeast / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationMiscellaneous transport and binding events / dolichol-linked oligosaccharide biosynthetic process / protein O-linked glycosylation via mannose / dolichyl-diphosphooligosaccharide-protein glycotransferase / dolichyl-diphosphooligosaccharide-protein glycotransferase activity / oligosaccharyltransferase complex / : / : / protein N-linked glycosylation / glycosyltransferase activity ...Miscellaneous transport and binding events / dolichol-linked oligosaccharide biosynthetic process / protein O-linked glycosylation via mannose / dolichyl-diphosphooligosaccharide-protein glycotransferase / dolichyl-diphosphooligosaccharide-protein glycotransferase activity / oligosaccharyltransferase complex / : / : / protein N-linked glycosylation / glycosyltransferase activity / protein-disulfide reductase activity / Neutrophil degranulation / post-translational protein modification / nuclear envelope / protein-macromolecule adaptor activity / endoplasmic reticulum membrane / structural molecule activity / endoplasmic reticulum / mitochondrion / metal ion binding / membrane Similarity search - Function | |||||||||
| Biological species |   | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.3 Å | |||||||||
 Authors Authors | Wild R / Kowal J | |||||||||
| Funding support |  Switzerland, 2 items Switzerland, 2 items
| |||||||||
 Citation Citation |  Journal: Science / Year: 2018 Journal: Science / Year: 2018Title: Structure of the yeast oligosaccharyltransferase complex gives insight into eukaryotic N-glycosylation. Authors: Rebekka Wild / Julia Kowal / Jillianne Eyring / Elsy M Ngwa / Markus Aebi / Kaspar P Locher /  Abstract: Oligosaccharyltransferase (OST) is an essential membrane protein complex in the endoplasmic reticulum, where it transfers an oligosaccharide from a dolichol-pyrophosphate-activated donor to ...Oligosaccharyltransferase (OST) is an essential membrane protein complex in the endoplasmic reticulum, where it transfers an oligosaccharide from a dolichol-pyrophosphate-activated donor to glycosylation sites of secretory proteins. Here we describe the atomic structure of yeast OST determined by cryo-electron microscopy, revealing a conserved subunit arrangement. The active site of the catalytic STT3 subunit points away from the center of the complex, allowing unhindered access to substrates. The dolichol-pyrophosphate moiety binds to a lipid-exposed groove of STT3, whereas two noncatalytic subunits and an ordered N-glycan form a membrane-proximal pocket for the oligosaccharide. The acceptor polypeptide site faces an oxidoreductase domain in stand-alone OST complexes or is immediately adjacent to the translocon, suggesting how eukaryotic OSTs efficiently glycosylate a large number of polypeptides before their folding. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_4161.map.gz emd_4161.map.gz | 62.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-4161-v30.xml emd-4161-v30.xml emd-4161.xml emd-4161.xml | 28.1 KB 28.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_4161_fsc.xml emd_4161_fsc.xml | 10.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_4161.png emd_4161.png | 50.2 KB | ||
| Filedesc metadata |  emd-4161.cif.gz emd-4161.cif.gz | 8 KB | ||
| Others |  emd_4161_additional_1.map.gz emd_4161_additional_1.map.gz emd_4161_additional_2.map.gz emd_4161_additional_2.map.gz | 7.6 MB 96.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-4161 http://ftp.pdbj.org/pub/emdb/structures/EMD-4161 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4161 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4161 | HTTPS FTP |
-Related structure data
| Related structure data |  6eznMC  4257C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_4161.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_4161.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | None | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.065 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: Map of OST complex in nanodisc, post-processed and masked
| File | emd_4161_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Map of OST complex in nanodisc, post-processed and masked | ||||||||||||
| Projections & Slices |
| ||||||||||||
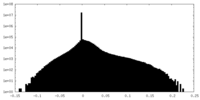
| Density Histograms |
-Additional map: Map of OST complex, after nanodisc subtraction, post-processed
| File | emd_4161_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Map of OST complex, after nanodisc subtraction, post-processed | ||||||||||||
| Projections & Slices |
| ||||||||||||
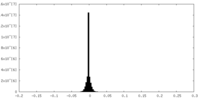
| Density Histograms |
- Sample components
Sample components
+Entire : yeast oligosaccharyltransferase (OST) complex
+Supramolecule #1: yeast oligosaccharyltransferase (OST) complex
+Macromolecule #1: Dolichyl-diphosphooligosaccharide--protein glycosyltransferase su...
+Macromolecule #2: Dolichyl-diphosphooligosaccharide--protein glycosyltransferase su...
+Macromolecule #3: Dolichyl-diphosphooligosaccharide--protein glycosyltransferase su...
+Macromolecule #4: Dolichyl-diphosphooligosaccharide--protein glycosyltransferase su...
+Macromolecule #5: Dolichyl-diphosphooligosaccharide--protein glycosyltransferase su...
+Macromolecule #6: Dolichyl-diphosphooligosaccharide--protein glycosyltransferase su...
+Macromolecule #7: Dolichyl-diphosphooligosaccharide--protein glycosyltransferase su...
+Macromolecule #8: Dolichyl-diphosphooligosaccharide--protein glycosyltransferase su...
+Macromolecule #12: 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE
+Macromolecule #13: PHOSPHATIDYLETHANOLAMINE
+Macromolecule #14: 2-acetamido-2-deoxy-beta-D-glucopyranose
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.5 mg/mL |
|---|---|
| Buffer | pH: 7.5 / Details: 20 mM HEPES, pH 7.5, 150 mM NaCl |
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 40 sec. |
| Vitrification | Cryogen name: ETHANE-PROPANE / Chamber humidity: 95 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV Details: Quantifoil carbon grids (300 mesh, copper) were glow discharged at 25 mA for 40 s. 3.5 ul of nanodisc-reconstituted OST sample at concentration of 0.5 mg/ml was applied onto the grid and ...Details: Quantifoil carbon grids (300 mesh, copper) were glow discharged at 25 mA for 40 s. 3.5 ul of nanodisc-reconstituted OST sample at concentration of 0.5 mg/ml was applied onto the grid and grids were blotted for 3 s and flash-frozen in a mixture of liquid ethane and propane cooled by liquid nitrogen.. |
| Details | OST complex reconstituted into nanodisc |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Quantum LS |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Number grids imaged: 2 / Number real images: 3915 / Average exposure time: 0.25 sec. / Average electron dose: 2.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-6ezn: |
 Movie
Movie Controller
Controller






















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)