[English] 日本語
 Yorodumi
Yorodumi- EMDB-3857: Structure of Aeropyrum pernix bacilliform virus 1 APBV1 helical capsid -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-3857 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of Aeropyrum pernix bacilliform virus 1 APBV1 helical capsid | |||||||||||||||
 Map data Map data | APBV1 helical capsid | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | archaeal virus / thermophile / capsid / helical / VIRUS | |||||||||||||||
| Function / homology | Major virion protein Function and homology information Function and homology information | |||||||||||||||
| Biological species |  Aeropyrum pernix bacilliform virus 1 Aeropyrum pernix bacilliform virus 1 | |||||||||||||||
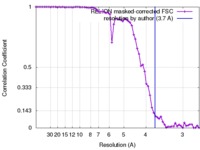
| Method | single particle reconstruction / cryo EM / Resolution: 3.7 Å | |||||||||||||||
 Authors Authors | Huiskonen JT / Ptchelkine D / Gillum A | |||||||||||||||
| Funding support |  United Kingdom, 4 items United Kingdom, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2017 Journal: Nat Commun / Year: 2017Title: Unique architecture of thermophilic archaeal virus APBV1 and its genome packaging. Authors: Denis Ptchelkine / Ashley Gillum / Tomohiro Mochizuki / Soizick Lucas-Staat / Ying Liu / Mart Krupovic / Simon E V Phillips / David Prangishvili / Juha T Huiskonen /     Abstract: Archaeal viruses have evolved to infect hosts often thriving in extreme conditions such as high temperatures. However, there is a paucity of information on archaeal virion structures, genome ...Archaeal viruses have evolved to infect hosts often thriving in extreme conditions such as high temperatures. However, there is a paucity of information on archaeal virion structures, genome packaging, and determinants of temperature resistance. The rod-shaped virus APBV1 (Aeropyrum pernix bacilliform virus 1) is among the most thermostable viruses known; it infects a hyperthermophile Aeropyrum pernix, which grows optimally at 90 °C. Here we report the structure of APBV1, determined by cryo-electron microscopy at near-atomic resolution. Tight packing of the major virion glycoprotein (VP1) is ensured by extended hydrophobic interfaces, and likely contributes to the extreme thermostability of the helical capsid. The double-stranded DNA is tightly packed in the capsid as a left-handed superhelix and held in place by the interactions with positively charged residues of VP1. The assembly is closed by specific capping structures at either end, which we propose to play a role in DNA packing and delivery. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_3857.map.gz emd_3857.map.gz | 20.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-3857-v30.xml emd-3857-v30.xml emd-3857.xml emd-3857.xml | 16.2 KB 16.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_3857_fsc.xml emd_3857_fsc.xml | 6.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_3857.png emd_3857.png | 344.2 KB | ||
| Masks |  emd_3857_msk_1.map emd_3857_msk_1.map | 22.2 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-3857.cif.gz emd-3857.cif.gz | 5.2 KB | ||
| Others |  emd_3857_half_map_1.map.gz emd_3857_half_map_1.map.gz emd_3857_half_map_2.map.gz emd_3857_half_map_2.map.gz | 20.4 MB 20.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-3857 http://ftp.pdbj.org/pub/emdb/structures/EMD-3857 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3857 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3857 | HTTPS FTP |
-Related structure data
| Related structure data |  5oxeMC  3858C  3859C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_3857.map.gz / Format: CCP4 / Size: 22.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_3857.map.gz / Format: CCP4 / Size: 22.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | APBV1 helical capsid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.35 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_3857_msk_1.map emd_3857_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: APBV1 helical capsid, half map 1
| File | emd_3857_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | APBV1 helical capsid, half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: APBV1 helical capsid, half map 2
| File | emd_3857_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | APBV1 helical capsid, half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Aeropyrum pernix bacilliform virus 1
| Entire | Name:  Aeropyrum pernix bacilliform virus 1 Aeropyrum pernix bacilliform virus 1 |
|---|---|
| Components |
|
-Supramolecule #1: Aeropyrum pernix bacilliform virus 1
| Supramolecule | Name: Aeropyrum pernix bacilliform virus 1 / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 700542 / Sci species name: Aeropyrum pernix bacilliform virus 1 / Virus type: VIRION / Virus isolate: SPECIES / Virus enveloped: No / Virus empty: No |
|---|---|
| Host (natural) | Organism:   Aeropyrum pernix (archaea) Aeropyrum pernix (archaea) |
-Macromolecule #1: Major virion protein
| Macromolecule | Name: Major virion protein / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Aeropyrum pernix bacilliform virus 1 Aeropyrum pernix bacilliform virus 1 |
| Molecular weight | Theoretical: 8.272858 KDa |
| Sequence | String: MAPKATLVKK FKGLAVGVGA LLAAPPIMGL ASYAVNGISS YLSITINSTT YDFAPLAQAV MVFGGIGLVA YGLHRILGRG L UniProtKB: Major virion protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | helical array |
- Sample preparation
Sample preparation
| Buffer | pH: 7 / Details: distilled water |
|---|---|
| Vitrification | Cryogen name: ETHANE-PROPANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI POLARA 300 |
|---|---|
| Specialist optics | Energy filter - Name: K2 Summit / Energy filter - Lower energy threshold: 0 eV / Energy filter - Upper energy threshold: 20 eV |
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Detector mode: COUNTING / Average exposure time: 0.2 sec. / Average electron dose: 1.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Calibrated magnification: 37037 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.0 mm |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-5oxe: |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)