[English] 日本語
 Yorodumi
Yorodumi- EMDB-30129: Helical stem of the cleaved double-headed nucleocapsids of sendai... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-30129 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Helical stem of the cleaved double-headed nucleocapsids of sendai virus | |||||||||
 Map data Map data | helical stem of cleaved double-headed nucleocapsids | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | nucleocapsid / Sendai virus / NUCLEAR PROTEIN / NUCLEAR PROTEIN-RNA complex | |||||||||
| Function / homology | Paramyxovirus nucleocapsid protein / Paramyxovirus nucleocapsid protein / helical viral capsid / viral nucleocapsid / host cell cytoplasm / ribonucleoprotein complex / structural molecule activity / RNA binding / Nucleoprotein Function and homology information Function and homology information | |||||||||
| Biological species |  Murine respirovirus / Murine respirovirus /  Sendai virus (strain Ohita) / synthetic construct (others) Sendai virus (strain Ohita) / synthetic construct (others) | |||||||||
| Method | helical reconstruction / cryo EM / Resolution: 2.9 Å | |||||||||
 Authors Authors | Shen Q / Shan H / Zhang N | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Commun Biol / Year: 2021 Journal: Commun Biol / Year: 2021Title: Structure and assembly of double-headed Sendai virus nucleocapsids. Authors: Na Zhang / Hong Shan / Mingdong Liu / Tianhao Li / Rui Luo / Liuyan Yang / Lei Qi / Xiaofeng Chu / Xin Su / Rui Wang / Yunhui Liu / Wenzhi Sun / Qing-Tao Shen /  Abstract: Paramyxoviruses, including the mumps virus, measles virus, Nipah virus and Sendai virus (SeV), have non-segmented single-stranded negative-sense RNA genomes which are encapsidated by nucleoproteins ...Paramyxoviruses, including the mumps virus, measles virus, Nipah virus and Sendai virus (SeV), have non-segmented single-stranded negative-sense RNA genomes which are encapsidated by nucleoproteins into helical nucleocapsids. Here, we reported a double-headed SeV nucleocapsid assembled in a tail-to-tail manner, and resolved its helical stems and clam-shaped joint at the respective resolutions of 2.9 and 3.9 Å, via cryo-electron microscopy. Our structures offer important insights into the mechanism of the helical polymerization, in particular via an unnoticed exchange of a N-terminal hole formed by three loops of nucleoproteins, and unveil the clam-shaped joint in a hyper-closed state for nucleocapsid dimerization. Direct visualization of the loop from the disordered C-terminal tail provides structural evidence that C-terminal tail is correlated to the curvature of nucleocapsid and links nucleocapsid condensation and genome replication and transcription with different assembly forms. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_30129.map.gz emd_30129.map.gz | 332.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-30129-v30.xml emd-30129-v30.xml emd-30129.xml emd-30129.xml | 12.9 KB 12.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_30129_fsc.xml emd_30129_fsc.xml | 18.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_30129.png emd_30129.png | 230.9 KB | ||
| Filedesc metadata |  emd-30129.cif.gz emd-30129.cif.gz | 5.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-30129 http://ftp.pdbj.org/pub/emdb/structures/EMD-30129 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30129 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30129 | HTTPS FTP |
-Related structure data
| Related structure data |  6m7dMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_30129.map.gz / Format: CCP4 / Size: 536.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_30129.map.gz / Format: CCP4 / Size: 536.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | helical stem of cleaved double-headed nucleocapsids | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.52 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
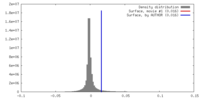
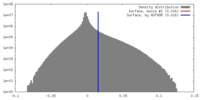
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : cleaved double-headed nucleocapsid of sendai virus
| Entire | Name: cleaved double-headed nucleocapsid of sendai virus |
|---|---|
| Components |
|
-Supramolecule #1: cleaved double-headed nucleocapsid of sendai virus
| Supramolecule | Name: cleaved double-headed nucleocapsid of sendai virus / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Murine respirovirus Murine respirovirus |
-Macromolecule #1: Nucleoprotein
| Macromolecule | Name: Nucleoprotein / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Sendai virus (strain Ohita) / Strain: Ohita Sendai virus (strain Ohita) / Strain: Ohita |
| Molecular weight | Theoretical: 57.20002 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MAGLLSTFDT FSSRRSESIN KSGGGAVIPG QRSTVSVFVL GPSVTDDADK LSIATTFLAH SLDTDKQHSQ RGGFLVSLLA MAYSSPELY LTTNGVNADV KYVIYNIEKD PKRTKTDGFI VKTRDMEYER TTEWLFGPMV NKSPLFQGQR DAADPDTLLQ I YGYPACLG ...String: MAGLLSTFDT FSSRRSESIN KSGGGAVIPG QRSTVSVFVL GPSVTDDADK LSIATTFLAH SLDTDKQHSQ RGGFLVSLLA MAYSSPELY LTTNGVNADV KYVIYNIEKD PKRTKTDGFI VKTRDMEYER TTEWLFGPMV NKSPLFQGQR DAADPDTLLQ I YGYPACLG AIIVQVWIVL VKAITSSAGL RKGFFNRLEA FRQDGTVKGA LVFTGETVEG IGSVMRSQQS LVSLMVETLV TM NTARSDL TTLEKNIQIV GNYIRDAGLA SFMNTIKYGV ETKMAALTLS NLRPDINKLR SLIDTYLSKG PRAPFICILK DPV HGEFAP GNYPALWSYA MGVAVVQNKA MQQYVTGRTY LDMEMFLLGQ AVAKDAESKI SSALEDELGV TDTAKERLRH HLAN LSGGD GAYHKPTGGG AIEVALDNAD IDLEPEAHTD QDARGWGGDS GDRWARSMGS GHFITLHGAE RLEEETNDED VSDIE RRIA RRLAERRQED ATTHEDEGRN NGVDHDEEDD AAAAAGMGGI UniProtKB: Nucleoprotein |
-Macromolecule #2: RNA (5'-R(P*UP*UP*UP*UP*UP*U)-3')
| Macromolecule | Name: RNA (5'-R(P*UP*UP*UP*UP*UP*U)-3') / type: rna / ID: 2 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 1.792037 KDa |
| Sequence | String: UUUUUU |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL | ||||||
|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 / Component:
| ||||||
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: CONTINUOUS | ||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 289 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average exposure time: 0.068 sec. / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)