[English] 日本語
 Yorodumi
Yorodumi- EMDB-3781: TET12SN density map (Design of in vivo self-assembling coiled-coi... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-3781 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | TET12SN density map (Design of in vivo self-assembling coiled-coil protein origami) | |||||||||
 Map data Map data | Bioorigami coiled-coil tetrahedron TET12SN. | |||||||||
 Sample Sample |
| |||||||||
| Biological species | Designed protein (others) | |||||||||
| Method | single particle reconstruction / negative staining / Resolution: 28.0 Å | |||||||||
 Authors Authors | Melero R / Carazo JM | |||||||||
 Citation Citation |  Journal: Nat Biotechnol / Year: 2017 Journal: Nat Biotechnol / Year: 2017Title: Design of coiled-coil protein-origami cages that self-assemble in vitro and in vivo. Authors: Ajasja Ljubetič / Fabio Lapenta / Helena Gradišar / Igor Drobnak / Jana Aupič / Žiga Strmšek / Duško Lainšček / Iva Hafner-Bratkovič / Andreja Majerle / Nuša Krivec / Mojca ...Authors: Ajasja Ljubetič / Fabio Lapenta / Helena Gradišar / Igor Drobnak / Jana Aupič / Žiga Strmšek / Duško Lainšček / Iva Hafner-Bratkovič / Andreja Majerle / Nuša Krivec / Mojca Benčina / Tomaž Pisanski / Tanja Ćirković Veličković / Adam Round / José María Carazo / Roberto Melero / Roman Jerala /      Abstract: Polypeptides and polynucleotides are natural programmable biopolymers that can self-assemble into complex tertiary structures. We describe a system analogous to designed DNA nanostructures in which ...Polypeptides and polynucleotides are natural programmable biopolymers that can self-assemble into complex tertiary structures. We describe a system analogous to designed DNA nanostructures in which protein coiled-coil (CC) dimers serve as building blocks for modular de novo design of polyhedral protein cages that efficiently self-assemble in vitro and in vivo. We produced and characterized >20 single-chain protein cages in three shapes-tetrahedron, four-sided pyramid, and triangular prism-with the largest containing >700 amino-acid residues and measuring 11 nm in diameter. Their stability and folding kinetics were similar to those of natural proteins. Solution small-angle X-ray scattering (SAXS), electron microscopy (EM), and biophysical analysis confirmed agreement of the expressed structures with the designs. We also demonstrated self-assembly of a tetrahedral structure in bacteria, mammalian cells, and mice without evidence of inflammation. A semi-automated computational design platform and a toolbox of CC building modules are provided to enable the design of protein cages in any polyhedral shape. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_3781.map.gz emd_3781.map.gz | 3.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-3781-v30.xml emd-3781-v30.xml emd-3781.xml emd-3781.xml | 9.8 KB 9.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_3781.png emd_3781.png | 46.2 KB | ||
| Filedesc metadata |  emd-3781.cif.gz emd-3781.cif.gz | 3.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-3781 http://ftp.pdbj.org/pub/emdb/structures/EMD-3781 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3781 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3781 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_3781.map.gz / Format: CCP4 / Size: 3.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_3781.map.gz / Format: CCP4 / Size: 3.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Bioorigami coiled-coil tetrahedron TET12SN. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.84 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : TET12SN
| Entire | Name: TET12SN |
|---|---|
| Components |
|
-Supramolecule #1: TET12SN
| Supramolecule | Name: TET12SN / type: complex / ID: 1 / Parent: 0 Details: Coiled coil single chain protein origami tetrahedron composed of soluble dimeric coiled coil segments. |
|---|---|
| Source (natural) | Organism: Designed protein (others) |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.03 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||||||||
| Staining | Type: NEGATIVE / Material: Uranyl Acetate Details: Samples were applied to glow discharged carbon-coated copper grids, washed quickly with distilled water and negatively stained with 2% (w/v) uranyl acetate. |
- Electron microscopy
Electron microscopy
| Microscope | JEOL 1230 |
|---|---|
| Image recording | Film or detector model: TVIPS TEMCAM-F416 (4k x 4k) / Average electron dose: 15.0 e/Å2 |
| Electron beam | Acceleration voltage: 100 kV / Electron source: TUNGSTEN HAIRPIN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm |
- Image processing
Image processing
| Startup model | Type of model: OTHER |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 28.0 Å / Resolution method: FSC 0.143 CUT-OFF / Software: (Name: Xmipp, RELION) / Number images used: 13425 |
| Initial angle assignment | Type: PROJECTION MATCHING |
| Final angle assignment | Type: PROJECTION MATCHING |
 Movie
Movie Controller
Controller


 UCSF Chimera
UCSF Chimera

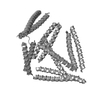

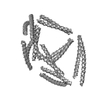



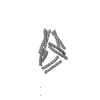













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















