[English] 日本語
 Yorodumi
Yorodumi- EMDB-3749: Poliovirus type 3 (strain Saukett) stabilized virus-like particle... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-3749 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Poliovirus type 3 (strain Saukett) stabilized virus-like particle in complex with the pocket factor compound GPP3 | |||||||||
 Map data Map data | MRC format map file. Contour level determined by visual inspection in UCSF Chimera. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Poliovirus / virus-like particle / vaccine / pocket factor / VIRUS LIKE PARTICLE | |||||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont genome entry into host cell via pore formation in plasma membrane / viral capsid / host cell cytoplasm / symbiont-mediated suppression of host gene expression / virion attachment to host cell / structural molecule activity Similarity search - Function | |||||||||
| Biological species |  Human poliovirus 3 Human poliovirus 3 | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.1 Å | |||||||||
 Authors Authors | Bahar MW / Kotecha A | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2017 Journal: Nat Commun / Year: 2017Title: Plant-made polio type 3 stabilized VLPs-a candidate synthetic polio vaccine. Authors: Johanna Marsian / Helen Fox / Mohammad W Bahar / Abhay Kotecha / Elizabeth E Fry / David I Stuart / Andrew J Macadam / David J Rowlands / George P Lomonossoff /  Abstract: Poliovirus (PV) is the causative agent of poliomyelitis, a crippling human disease known since antiquity. PV occurs in two distinct antigenic forms, D and C, of which only the D form elicits a robust ...Poliovirus (PV) is the causative agent of poliomyelitis, a crippling human disease known since antiquity. PV occurs in two distinct antigenic forms, D and C, of which only the D form elicits a robust neutralizing response. Developing a synthetically produced stabilized virus-like particle (sVLP)-based vaccine with D antigenicity, without the drawbacks of current vaccines, will be a major step towards the final eradication of poliovirus. Such a sVLP would retain the native antigenic conformation and the repetitive structure of the original virus particle, but lack infectious genomic material. In this study, we report the production of synthetically stabilized PV VLPs in plants. Mice carrying the gene for the human PV receptor are protected from wild-type PV when immunized with the plant-made PV sVLPs. Structural analysis of the stabilized mutant at 3.6 Å resolution by cryo-electron microscopy and single-particle reconstruction reveals a structure almost indistinguishable from wild-type PV3.Despite the success of current vaccination against poliomyelitis, safe, cheap and effective vaccines remain sought for continuing eradication effort. Here the authors use plants to express stabilized virus-like particles of type 3 poliovirus that can induce a protective immune response in mice transgenic for the human poliovirus receptor. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_3749.map.gz emd_3749.map.gz | 480.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-3749-v30.xml emd-3749-v30.xml emd-3749.xml emd-3749.xml | 19.7 KB 19.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_3749_fsc.xml emd_3749_fsc.xml | 17.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_3749.png emd_3749.png | 326.4 KB | ||
| Filedesc metadata |  emd-3749.cif.gz emd-3749.cif.gz | 7.1 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-3749 http://ftp.pdbj.org/pub/emdb/structures/EMD-3749 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3749 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3749 | HTTPS FTP |
-Related structure data
| Related structure data |  5o5pMC  3747C  5o5bC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_3749.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_3749.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | MRC format map file. Contour level determined by visual inspection in UCSF Chimera. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.35 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Human poliovirus 3
| Entire | Name:  Human poliovirus 3 Human poliovirus 3 |
|---|---|
| Components |
|
-Supramolecule #1: Human poliovirus 3
| Supramolecule | Name: Human poliovirus 3 / type: virus / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 Details: Poliovirus type 3 (Saukett strain) virus-like particle produced in plant expression system. NCBI-ID: 12086 / Sci species name: Human poliovirus 3 / Sci species strain: Saukett / Virus type: VIRUS-LIKE PARTICLE / Virus isolate: SEROTYPE / Virus enveloped: No / Virus empty: Yes |
|---|---|
| Host (natural) | Organism:  Human poliovirus 3 / Strain: Saukett Human poliovirus 3 / Strain: Saukett |
| Molecular weight | Theoretical: 5.8 MDa |
| Virus shell | Shell ID: 1 / Name: Capsid / Diameter: 327.0 Å / T number (triangulation number): 1 |
-Macromolecule #1: Capsid proteins, VP1
| Macromolecule | Name: Capsid proteins, VP1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Human poliovirus 3 / Strain: Saukett Human poliovirus 3 / Strain: Saukett |
| Molecular weight | Theoretical: 33.562785 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GIEDLITEVA QGALTLSLPK QQDSLPDTKA SGPAHSKEVP ALTAVETGAT NPLVPSDTVQ TRHVIQRRSR SESTIESFFA RGACVAIIE VDNEEPTTRA QKLFAMWRIT YKDTVQLRRK LEFFTYSRFD MELTFVVTAN FTNTNNGHAL NQVYQIMYIP P GAPTPKSW ...String: GIEDLITEVA QGALTLSLPK QQDSLPDTKA SGPAHSKEVP ALTAVETGAT NPLVPSDTVQ TRHVIQRRSR SESTIESFFA RGACVAIIE VDNEEPTTRA QKLFAMWRIT YKDTVQLRRK LEFFTYSRFD MELTFVVTAN FTNTNNGHAL NQVYQIMYIP P GAPTPKSW DDYTWQTSSN PSIFYTYGAA PARISVPYVG LANAYSHFYD GFAKVPLKTD ANDQIGDSLY SAMTVDDFGV LA IRVVNDH NPTKVTSKVR IYMKPKHVRV WCPRPPRAVP YYGPGVDYKD NLNPLSEKGL TTY UniProtKB: Genome polyprotein |
-Macromolecule #2: Capsid proteins, VP2
| Macromolecule | Name: Capsid proteins, VP2 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Human poliovirus 3 / Strain: Saukett Human poliovirus 3 / Strain: Saukett |
| Molecular weight | Theoretical: 30.188982 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: SPNVEACGYS DRVLQLTIGN STITTQEAAN SVVAYGRWPE FIRDDEANPV DQPTEPDVAT CRFYTLDTVM WGKESKGWWW KLPDALRDM GLFGQNMYYH YLGRSGYTVH VQCNASKFHQ GALGVFAIPE YCLAGDSDKQ RYTSYANANP GEKGGKFYSQ F NRDTAVTS ...String: SPNVEACGYS DRVLQLTIGN STITTQEAAN SVVAYGRWPE FIRDDEANPV DQPTEPDVAT CRFYTLDTVM WGKESKGWWW KLPDALRDM GLFGQNMYYH YLGRSGYTVH VQCNASKFHQ GALGVFAIPE YCLAGDSDKQ RYTSYANANP GEKGGKFYSQ F NRDTAVTS PKREFCPVDY LLGCGVLLGN AFVYPHQIIN LRTNNSATIV LPYVNAMAID SMVKHNNWGI AILPLSPLDF AQ ESSVEIP ITVTIAPMCS EFNGLRNVTA PKFQ UniProtKB: Genome polyprotein |
-Macromolecule #3: Capsid proteins, VP3
| Macromolecule | Name: Capsid proteins, VP3 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Human poliovirus 3 / Strain: Saukett Human poliovirus 3 / Strain: Saukett |
| Molecular weight | Theoretical: 26.3151 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GLPVLNTPGS NQYLTSDNYQ SPCAIPEFDV TPPIDIPGEV KNMMELAEID TMIPLNLENT KRNTMDMYRV TLSDSADLSQ PILCFSLSP ASDPRLSHTM LGEVLNYYTH WAGSLKFTFL FCGSMMATGK ILVAYAPPGA QPPTSRKEAM LGTHVIWDLG L QSSCTMVV ...String: GLPVLNTPGS NQYLTSDNYQ SPCAIPEFDV TPPIDIPGEV KNMMELAEID TMIPLNLENT KRNTMDMYRV TLSDSADLSQ PILCFSLSP ASDPRLSHTM LGEVLNYYTH WAGSLKFTFL FCGSMMATGK ILVAYAPPGA QPPTSRKEAM LGTHVIWDLG L QSSCTMVV PWISNVTYRQ TTQDSFTEGG YISMFYQTRI VVPLSTPKSM SMLGFVSACN DFSVRLLRDT THISQSALPQ UniProtKB: Genome polyprotein |
-Macromolecule #4: Capsid proteins, VP4
| Macromolecule | Name: Capsid proteins, VP4 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Human poliovirus 3 / Strain: Saukett Human poliovirus 3 / Strain: Saukett |
| Molecular weight | Theoretical: 7.452113 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGAQVSSQKV GAHENSNRAY GGSTINYTTI NYYKDSASNA ASKQDYSQDP SKFTEPLKDV LIKTAPALN UniProtKB: Genome polyprotein |
-Macromolecule #5: 1-[5-[4-(ethoxyiminomethyl)phenoxy]-3-methyl-pentyl]-3-pyridin-4-...
| Macromolecule | Name: 1-[5-[4-(ethoxyiminomethyl)phenoxy]-3-methyl-pentyl]-3-pyridin-4-yl-imidazol-2-one type: ligand / ID: 5 / Number of copies: 1 / Formula: 9LW |
|---|---|
| Molecular weight | Theoretical: 408.493 Da |
| Chemical component information | 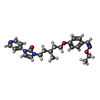 ChemComp-9LW: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.1 mg/mL |
|---|---|
| Buffer | pH: 7 / Details: PBS pH 7.0 |
| Grid | Model: C-flat CF-2/1-2C / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 293 K / Instrument: FEI VITROBOT MARK I / Details: Blot for 4 seconds before plunging.. |
| Details | Virus-like particles for polio type 3 (strain Saukett) in complex with pocket factor compound GPP3 were assessed by negative stain EM analysis, prior to cryo-em data collection. |
- Electron microscopy
Electron microscopy
| Microscope | FEI POLARA 300 |
|---|---|
| Specialist optics | Energy filter - Name: GIF Quantum / Energy filter - Lower energy threshold: 0 eV / Energy filter - Upper energy threshold: 20 eV |
| Details | Preliminary grid screening was performed. |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 4086 pixel / Digitization - Dimensions - Height: 4086 pixel / Digitization - Frames/image: 2-25 / Number grids imaged: 1 / Number real images: 1018 / Average exposure time: 0.2 sec. / Average electron dose: 1.2 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Calibrated magnification: 37037 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.0 mm / Nominal defocus max: 2.8000000000000003 µm / Nominal defocus min: 0.8 µm |
| Sample stage | Specimen holder model: GATAN 910 MULTI-SPECIMEN SINGLE TILT CRYO TRANSFER HOLDER Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Details | phenix.real_space_refine was used to refine the atomic model in the cryo-em map. |
|---|---|
| Refinement | Space: REAL / Protocol: RIGID BODY FIT / Target criteria: Cross-correlation coefficient |
| Output model |  PDB-5o5p: |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)






















