[English] 日本語
 Yorodumi
Yorodumi- EMDB-4098: Cryo-EM reconstruction of bacteriophage AP205 virus-like particles -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-4098 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM reconstruction of bacteriophage AP205 virus-like particles | |||||||||
 Map data Map data | Bacteriophage AP205 cryo-EM map | |||||||||
 Sample Sample | AP205 != Acinetobacter phage AP205 AP205
| |||||||||
 Keywords Keywords | RNA bacteriophage / Leviviridae / coat protein / virus-like particle / VIRUS LIKE PARTICLE | |||||||||
| Function / homology | : / AP205 coat protein / viral capsid / Coat protein Function and homology information Function and homology information | |||||||||
| Biological species |  Acinetobacter phage AP205 (virus) Acinetobacter phage AP205 (virus) | |||||||||
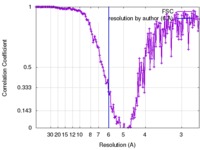
| Method | single particle reconstruction / cryo EM / Resolution: 6.0 Å | |||||||||
 Authors Authors | Diebolder CA / Rumnieks J | |||||||||
 Citation Citation |  Journal: J Mol Biol / Year: 2016 Journal: J Mol Biol / Year: 2016Title: Structure of AP205 Coat Protein Reveals Circular Permutation in ssRNA Bacteriophages. Authors: Mihails Shishovs / Janis Rumnieks / Christoph Diebolder / Kristaps Jaudzems / Loren B Andreas / Jan Stanek / Andris Kazaks / Svetlana Kotelovica / Inara Akopjana / Guido Pintacuda / Roman I ...Authors: Mihails Shishovs / Janis Rumnieks / Christoph Diebolder / Kristaps Jaudzems / Loren B Andreas / Jan Stanek / Andris Kazaks / Svetlana Kotelovica / Inara Akopjana / Guido Pintacuda / Roman I Koning / Kaspars Tars /    Abstract: AP205 is a single-stranded RNA bacteriophage that has a coat protein sequence not similar to any other known single-stranded RNA phage. Here, we report an atomic-resolution model of the AP205 virus- ...AP205 is a single-stranded RNA bacteriophage that has a coat protein sequence not similar to any other known single-stranded RNA phage. Here, we report an atomic-resolution model of the AP205 virus-like particle based on a crystal structure of an unassembled coat protein dimer and a cryo-electron microscopy reconstruction of the assembled particle, together with secondary structure information from site-specific solid-state NMR data. The AP205 coat protein dimer adopts the conserved Leviviridae coat protein fold except for the N-terminal region, which forms a beta-hairpin in the other known single-stranded RNA phages. AP205 has a similar structure at the same location formed by N- and C-terminal beta-strands, making it a circular permutant compared to the other coat proteins. The permutation moves the coat protein termini to the most surface-exposed part of the assembled particle, which explains its increased tolerance to long N- and C-terminal fusions. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_4098.map.gz emd_4098.map.gz | 226.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-4098-v30.xml emd-4098-v30.xml emd-4098.xml emd-4098.xml | 12.1 KB 12.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_4098_fsc.xml emd_4098_fsc.xml | 16.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_4098.png emd_4098.png | 225.9 KB | ||
| Filedesc metadata |  emd-4098.cif.gz emd-4098.cif.gz | 5.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-4098 http://ftp.pdbj.org/pub/emdb/structures/EMD-4098 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4098 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4098 | HTTPS FTP |
-Related structure data
| Related structure data |  5lqpMC  5fs4C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_4098.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_4098.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Bacteriophage AP205 cryo-EM map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.34 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : AP205
| Entire | Name: AP205 |
|---|---|
| Components |
|
-Supramolecule #1: Acinetobacter phage AP205
| Supramolecule | Name: Acinetobacter phage AP205 / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 154784 / Sci species name: Acinetobacter phage AP205 / Virus type: VIRUS-LIKE PARTICLE / Virus isolate: SPECIES / Virus enveloped: No / Virus empty: No |
|---|
-Macromolecule #1: Coat protein
| Macromolecule | Name: Coat protein / type: protein_or_peptide / ID: 1 / Number of copies: 180 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Acinetobacter phage AP205 (virus) Acinetobacter phage AP205 (virus) |
| Molecular weight | Theoretical: 13.820569 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: ANKPMQPITS TANKIVWSDP TRLSTTFSAS LLRQRVKVGI AELNNVSGQY VSVYKRPAPK PEGCADACVI MPNENQSIRT VISGSAENL ATLKAEWETH KRNVDTLFAS GNAGLGFLDP TAAIVSSDTT UniProtKB: Coat protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 400 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 298 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: FEI SFEG |
| Image recording | Film or detector model: FEI FALCON II (4k x 4k) / Detector mode: INTEGRATING / Digitization - Frames/image: 1-7 / Average electron dose: 8.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated defocus max: 2.5 µm / Calibrated defocus min: 1.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 59000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)