+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-3675 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Nucleosome core particle Ubiquitylated at H2A Lys-13 | |||||||||
 Map data Map data | Nucleosome core particle with ubquitin covalently tethered to H2A lysine 13 | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
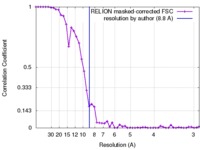
| Method | single particle reconstruction / cryo EM / Resolution: 8.8 Å | |||||||||
 Authors Authors | Wilson MD / Kitevski-LeBlanc J / Durocher D / Rubinstein JL / Kay LE | |||||||||
 Citation Citation |  Journal: Elife / Year: 2017 Journal: Elife / Year: 2017Title: The RNF168 paralog RNF169 defines a new class of ubiquitylated histone reader involved in the response to DNA damage. Authors: Julianne Kitevski-LeBlanc / Amélie Fradet-Turcotte / Predrag Kukic / Marcus D Wilson / Guillem Portella / Tairan Yuwen / Stephanie Panier / Shili Duan / Marella D Canny / Hugo van Ingen / ...Authors: Julianne Kitevski-LeBlanc / Amélie Fradet-Turcotte / Predrag Kukic / Marcus D Wilson / Guillem Portella / Tairan Yuwen / Stephanie Panier / Shili Duan / Marella D Canny / Hugo van Ingen / Cheryl H Arrowsmith / John L Rubinstein / Michele Vendruscolo / Daniel Durocher / Lewis E Kay /    Abstract: Site-specific histone ubiquitylation plays a central role in orchestrating the response to DNA double-strand breaks (DSBs). DSBs elicit a cascade of events controlled by the ubiquitin ligase RNF168, ...Site-specific histone ubiquitylation plays a central role in orchestrating the response to DNA double-strand breaks (DSBs). DSBs elicit a cascade of events controlled by the ubiquitin ligase RNF168, which promotes the accumulation of repair factors such as 53BP1 and BRCA1 on the chromatin flanking the break site. RNF168 also promotes its own accumulation, and that of its paralog RNF169, but how they recognize ubiquitylated chromatin is unknown. Using methyl-TROSY solution NMR spectroscopy and molecular dynamics simulations, we present an atomic resolution model of human RNF169 binding to a ubiquitylated nucleosome, and validate it by electron cryomicroscopy. We establish that RNF169 binds to ubiquitylated H2A-Lys13/Lys15 in a manner that involves its canonical ubiquitin-binding helix and a pair of arginine-rich motifs that interact with the nucleosome acidic patch. This three-pronged interaction mechanism is distinct from that by which 53BP1 binds to ubiquitylated H2A-Lys15 highlighting the diversity in site-specific recognition of ubiquitylated nucleosomes. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_3675.map.gz emd_3675.map.gz | 3.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-3675-v30.xml emd-3675-v30.xml emd-3675.xml emd-3675.xml | 12.4 KB 12.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_3675_fsc.xml emd_3675_fsc.xml | 4.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_3675.png emd_3675.png | 38 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-3675 http://ftp.pdbj.org/pub/emdb/structures/EMD-3675 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3675 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3675 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_3675.map.gz / Format: CCP4 / Size: 8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_3675.map.gz / Format: CCP4 / Size: 8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Nucleosome core particle with ubquitin covalently tethered to H2A lysine 13 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.45 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : NCP-ub complex
| Entire | Name: NCP-ub complex |
|---|---|
| Components |
|
-Supramolecule #1: NCP-ub complex
| Supramolecule | Name: NCP-ub complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 Details: H2AK13 disulphide ubiquitylated Nucleosome core particle |
|---|
-Supramolecule #2: H2AK13 ubiquitylated Nucleosome core particle
| Supramolecule | Name: H2AK13 ubiquitylated Nucleosome core particle / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 Details: recombinant Drosophila histones wrapped with synthetic strong positioning Widom-601 DNA. Ubiquitin G76C covalently tethered to engineered K13C residue in H2A, prior to NCP reconstitution |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
| Molecular weight | Theoretical: 220 kDa/nm |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.65 mg/mL |
|---|---|
| Buffer | pH: 7.5 / Details: 10 mM Tris-Cl 17 pH 7.5, 50 mM KCl, 1 mM EDTA |
| Grid | Model: homemade / Material: GOLD / Mesh: 400 / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE-PROPANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK III |
| Details | NCP-ub |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 1 / Number real images: 175 / Average exposure time: 15.0 sec. / Average electron dose: 36.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 34483 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.0 mm / Nominal magnification: 25000 |
| Sample stage | Specimen holder model: GATAN 626 SINGLE TILT LIQUID NITROGEN CRYO TRANSFER HOLDER Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)