[English] 日本語
 Yorodumi
Yorodumi- EMDB-10240: Structure of the native full-length HIV-1 capsid protein A92E in ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10240 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the native full-length HIV-1 capsid protein A92E in helical assembly (-13,11) | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | HIV / capsid / hexamer / helical assembly / curvature / VIRAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationintegrase activity / Integration of viral DNA into host genomic DNA / Autointegration results in viral DNA circles / Minus-strand DNA synthesis / Plus-strand DNA synthesis / Uncoating of the HIV Virion / 2-LTR circle formation / Vpr-mediated nuclear import of PICs / Early Phase of HIV Life Cycle / Integration of provirus ...integrase activity / Integration of viral DNA into host genomic DNA / Autointegration results in viral DNA circles / Minus-strand DNA synthesis / Plus-strand DNA synthesis / Uncoating of the HIV Virion / 2-LTR circle formation / Vpr-mediated nuclear import of PICs / Early Phase of HIV Life Cycle / Integration of provirus / APOBEC3G mediated resistance to HIV-1 infection / Binding and entry of HIV virion / viral life cycle / HIV-1 retropepsin / symbiont-mediated activation of host apoptosis / retroviral ribonuclease H / exoribonuclease H / exoribonuclease H activity / Assembly Of The HIV Virion / protein processing / Budding and maturation of HIV virion / viral genome integration into host DNA / establishment of integrated proviral latency / RNA-directed DNA polymerase / RNA stem-loop binding / viral penetration into host nucleus / host multivesicular body / RNA-directed DNA polymerase activity / RNA-DNA hybrid ribonuclease activity / Transferases; Transferring phosphorus-containing groups; Nucleotidyltransferases / peptidase activity / host cell / viral nucleocapsid / DNA recombination / DNA-directed DNA polymerase / aspartic-type endopeptidase activity / Hydrolases; Acting on ester bonds / host cell cytoplasm / DNA-directed DNA polymerase activity / symbiont-mediated suppression of host gene expression / viral translational frameshifting / symbiont entry into host cell / lipid binding / host cell nucleus / host cell plasma membrane / virion membrane / structural molecule activity / DNA binding / RNA binding / zinc ion binding / identical protein binding / membrane Similarity search - Function | |||||||||
| Biological species |   Human immunodeficiency virus 1 Human immunodeficiency virus 1 | |||||||||
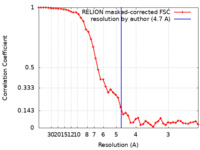
| Method | helical reconstruction / cryo EM / Resolution: 4.7 Å | |||||||||
 Authors Authors | Ni T / Gerard S | |||||||||
| Funding support |  United Kingdom, United Kingdom,  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2020 Journal: Nat Struct Mol Biol / Year: 2020Title: Intrinsic curvature of the HIV-1 CA hexamer underlies capsid topology and interaction with cyclophilin A. Authors: Tao Ni / Samuel Gerard / Gongpu Zhao / Kyle Dent / Jiying Ning / Jing Zhou / Jiong Shi / Jordan Anderson-Daniels / Wen Li / Sooin Jang / Alan N Engelman / Christopher Aiken / Peijun Zhang /   Abstract: The mature retrovirus capsid consists of a variably curved lattice of capsid protein (CA) hexamers and pentamers. High-resolution structures of the curved assembly, or in complex with host factors, ...The mature retrovirus capsid consists of a variably curved lattice of capsid protein (CA) hexamers and pentamers. High-resolution structures of the curved assembly, or in complex with host factors, have not been available. By devising cryo-EM methodologies for exceedingly flexible and pleomorphic assemblies, we have determined cryo-EM structures of apo-CA hexamers and in complex with cyclophilin A (CypA) at near-atomic resolutions. The CA hexamers are intrinsically curved, flexible and asymmetric, revealing the capsomere and not the previously touted dimer or trimer interfaces as the key contributor to capsid curvature. CypA recognizes specific geometries of the curved lattice, simultaneously interacting with three CA protomers from adjacent hexamers via two noncanonical interfaces, thus stabilizing the capsid. By determining multiple structures from various helical symmetries, we further revealed the essential plasticity of the CA molecule, which allows formation of continuously curved conical capsids and the mechanism of capsid pattern sensing by CypA. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10240.map.gz emd_10240.map.gz | 1.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10240-v30.xml emd-10240-v30.xml emd-10240.xml emd-10240.xml | 16.2 KB 16.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_10240_fsc.xml emd_10240_fsc.xml | 4.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_10240.png emd_10240.png | 86.2 KB | ||
| Masks |  emd_10240_msk_1.map emd_10240_msk_1.map | 8 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-10240.cif.gz emd-10240.cif.gz | 5.7 KB | ||
| Others |  emd_10240_half_map_1.map.gz emd_10240_half_map_1.map.gz emd_10240_half_map_2.map.gz emd_10240_half_map_2.map.gz | 5.4 MB 5.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10240 http://ftp.pdbj.org/pub/emdb/structures/EMD-10240 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10240 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10240 | HTTPS FTP |
-Related structure data
| Related structure data |  6sluMC  6skkC  6skmC  6sknC  6slqC  6smuC  6y9vC  6y9wC  6y9xC  6y9yC  6y9zC  6yj5C  6zdjC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_10240.map.gz / Format: CCP4 / Size: 8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_10240.map.gz / Format: CCP4 / Size: 8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.22 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_10240_msk_1.map emd_10240_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_10240_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_10240_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : In vitro assembled HIV-1 capsid in tubular assembly
| Entire | Name: In vitro assembled HIV-1 capsid in tubular assembly |
|---|---|
| Components |
|
-Supramolecule #1: In vitro assembled HIV-1 capsid in tubular assembly
| Supramolecule | Name: In vitro assembled HIV-1 capsid in tubular assembly / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
-Macromolecule #1: Gag protein
| Macromolecule | Name: Gag protein / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Molecular weight | Theoretical: 25.66442 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: PIVQNIQGQM VHQAISPRTL NAWVKVVEEK AFSPEVIPMF SALSEGATPQ DLNTMLNTVG GHQAAMQMLK ETINEEAAEW DRVHPVHAG PIEPGQMREP RGSDIAGTTS TLQEQIGWMT NNPPIPVGEI YKRWIILGLN KIVRMYSPTS ILDIRQGPKE P FRDYVDRF ...String: PIVQNIQGQM VHQAISPRTL NAWVKVVEEK AFSPEVIPMF SALSEGATPQ DLNTMLNTVG GHQAAMQMLK ETINEEAAEW DRVHPVHAG PIEPGQMREP RGSDIAGTTS TLQEQIGWMT NNPPIPVGEI YKRWIILGLN KIVRMYSPTS ILDIRQGPKE P FRDYVDRF YKTLRAEQAS QEVKNWMTET LLVQNANPDC KTILKALGPA ATLEEMMTAC QGVGGPGHKA RVL UniProtKB: Gag protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | helical array |
- Sample preparation
Sample preparation
| Concentration | 2 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Vitrification | Cryogen name: ETHANE |
| Details | Purified capsid protein self-assembled in 50 mM Tris-HCl pH8.0 1M NaCl |
- Electron microscopy
Electron microscopy
| Microscope | FEI POLARA 300 |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Number real images: 6797 / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.0 mm |
| Sample stage | Specimen holder model: OTHER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
- Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT / Overall B value: 72.14 / Target criteria: Cross-correlation coefficient |
|---|---|
| Output model |  PDB-6slu: |
 Movie
Movie Controller
Controller































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)