[English] 日本語
 Yorodumi
Yorodumi- PDB-4j8v: X-ray structure of NCP145 with bound chlorido(eta-6-p-cymene)(N-p... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 4j8v | ||||||
|---|---|---|---|---|---|---|---|
| Title | X-ray structure of NCP145 with bound chlorido(eta-6-p-cymene)(N-phenyl-2-pyridinecarbothioamide)ruthenium(II) | ||||||
 Components Components |
| ||||||
 Keywords Keywords | STRUCTURAL PROTEIN/DNA / nucleosome / histone / STRUCTURAL PROTEIN-DNA complex | ||||||
| Function / homology |  Function and homology information Function and homology informationstructural constituent of chromatin / heterochromatin formation / nucleosome / nucleosome assembly / protein heterodimerization activity / DNA binding / nucleoplasm / nucleus Similarity search - Function | ||||||
| Biological species | synthetic construct (others) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.58 Å MOLECULAR REPLACEMENT / Resolution: 2.58 Å | ||||||
 Authors Authors | Adhireksan, Z. / Davey, C.A. | ||||||
 Citation Citation |  Journal: CHEM SCI / Year: 2013 Journal: CHEM SCI / Year: 2013Title: Novel metal(II) arene 2-pyridinecarbothioamides: a rationale to orally active organometallic anticancer agents Authors: Meier, S.M. / Hanif, M. / Adhireksan, Z. / Pichler, V. / Novak, M. / Jirkovsky, E. / Jakupec, M.A. / Arion, V.B. / Davey, C.A. / Keppler, B.K. / Hartinger, C.G. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  4j8v.cif.gz 4j8v.cif.gz | 320.9 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb4j8v.ent.gz pdb4j8v.ent.gz | 243.7 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  4j8v.json.gz 4j8v.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/j8/4j8v https://data.pdbj.org/pub/pdb/validation_reports/j8/4j8v ftp://data.pdbj.org/pub/pdb/validation_reports/j8/4j8v ftp://data.pdbj.org/pub/pdb/validation_reports/j8/4j8v | HTTPS FTP |
|---|
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| 2 | 
| ||||||||
| 3 |
| ||||||||
| Unit cell |
|
- Components
Components
-Protein , 4 types, 8 molecules AEBFCGDH
| #1: Protein | Mass: 15303.930 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  #2: Protein | Mass: 11263.231 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  #3: Protein | Mass: 13907.163 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  #4: Protein | Mass: 13848.097 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  |
|---|
-DNA chain , 2 types, 2 molecules IJ
| #5: DNA chain | Mass: 44749.664 Da / Num. of mol.: 1 / Source method: obtained synthetically / Source: (synth.) synthetic construct (others) |
|---|---|
| #6: DNA chain | Mass: 44740.648 Da / Num. of mol.: 1 / Source method: obtained synthetically / Source: (synth.) synthetic construct (others) |
-Non-polymers , 3 types, 6 molecules 
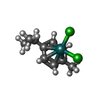



| #7: Chemical | | #8: Chemical | #9: Chemical | ChemComp-MG / | |
|---|
-Details
| Compound details | CHLORIDO(ETA-6-P-CYMENE)(N-PHENYL-2-PYRIDINECARBOTHIOAMIDE)RUTHENIUM(II) WAS USED IN ...CHLORIDO(ETA-6-P-CYMENE)(N-PHENYL-2-PYRIDINECA |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.7 Å3/Da / Density % sol: 54.42 % |
|---|---|
| Crystal grow | Temperature: 291 K / Method: vapor diffusion, hanging drop / pH: 6 Details: 40 mM MnCl2, 30 mM KCl, 20 mM K-Cacodylate pH 6.0, VAPOR DIFFUSION, HANGING DROP, temperature 291K |
-Data collection
| Diffraction | Mean temperature: 90 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  SLS SLS  / Beamline: X06DA / Wavelength: 1.5 Å / Beamline: X06DA / Wavelength: 1.5 Å |
| Detector | Type: MARMOSAIC 225 mm CCD / Detector: CCD / Date: Jul 3, 2011 |
| Radiation | Monochromator: Bartels Monochromator / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1.5 Å / Relative weight: 1 |
| Reflection | Resolution: 2.58→94.07 Å / Num. obs: 57178 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT / Resolution: 2.58→94.07 Å / Cor.coef. Fo:Fc: 0.925 / Cor.coef. Fo:Fc free: 0.898 / SU B: 11.367 / SU ML: 0.256 / Cross valid method: THROUGHOUT / ESU R: 0.869 / ESU R Free: 0.35 / Stereochemistry target values: MAXIMUM LIKELIHOOD / Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS MOLECULAR REPLACEMENT / Resolution: 2.58→94.07 Å / Cor.coef. Fo:Fc: 0.925 / Cor.coef. Fo:Fc free: 0.898 / SU B: 11.367 / SU ML: 0.256 / Cross valid method: THROUGHOUT / ESU R: 0.869 / ESU R Free: 0.35 / Stereochemistry target values: MAXIMUM LIKELIHOOD / Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.4 Å / Solvent model: MASK | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 77.273 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.58→94.07 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 2.58→2.647 Å / Total num. of bins used: 20
|
 Movie
Movie Controller
Controller















 PDBj
PDBj









































