+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-9802 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Nucleosome bound to C-terminal ELYS fragment | |||||||||
 Map data Map data | The structure of the nucleosome bound to C-terminal ELYS fragment. | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
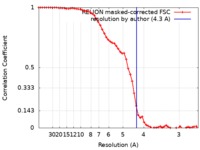
| Method | single particle reconstruction / cryo EM / Resolution: 4.3 Å | |||||||||
 Authors Authors | Kobayashi W / Takizawa Y / Aihara M / Negishi L / Ishii H / Kurumizaka H | |||||||||
 Citation Citation |  Journal: Commun Biol / Year: 2019 Journal: Commun Biol / Year: 2019Title: Structural and biochemical analyses of the nuclear pore complex component ELYS identify residues responsible for nucleosome binding. Authors: Wataru Kobayashi / Yoshimasa Takizawa / Maya Aihara / Lumi Negishi / Hajime Ishii / Hitoshi Kurumizaka /  Abstract: The nuclear pore complex embedded within the nuclear envelope is the essential architecture for trafficking macromolecules, such as proteins and RNAs, between the cytoplasm and nucleus. The nuclear ...The nuclear pore complex embedded within the nuclear envelope is the essential architecture for trafficking macromolecules, such as proteins and RNAs, between the cytoplasm and nucleus. The nuclear pore complex assembly occurs on chromatin in the post-mitotic phase of the cell cycle. ELYS (MEL-28/AHCTF1) binds to the nucleosome, which is the basic chromatin unit, and promotes assembly of the complex around the chromosomes in cells. Here we show that the Arg-Arg-Lys (RRK) stretch of the C-terminal ELYS region plays an essential role in the nucleosome binding. The cryo-EM structure and the crosslinking mass spectrometry reveal that the ELYS C-terminal region directly binds to the acidic patch of the nucleosome. These results provide mechanistic insight into the ELYS-nucleosome interaction, which promotes the post-mitotic nuclear pore complex formation around chromosomes in cells. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_9802.map.gz emd_9802.map.gz | 1.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-9802-v30.xml emd-9802-v30.xml emd-9802.xml emd-9802.xml | 10.3 KB 10.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_9802_fsc.xml emd_9802_fsc.xml | 5.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_9802.png emd_9802.png | 91.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-9802 http://ftp.pdbj.org/pub/emdb/structures/EMD-9802 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9802 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9802 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_9802.map.gz / Format: CCP4 / Size: 15.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_9802.map.gz / Format: CCP4 / Size: 15.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | The structure of the nucleosome bound to C-terminal ELYS fragment. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.32 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Nucleosome bound to C-terminal ELYS fragment
| Entire | Name: Nucleosome bound to C-terminal ELYS fragment |
|---|---|
| Components |
|
-Supramolecule #1: Nucleosome bound to C-terminal ELYS fragment
| Supramolecule | Name: Nucleosome bound to C-terminal ELYS fragment / type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Recombinant plasmid: pGEX6P-1-xELYS(2299-2408), pET15b-H3.1, pET15b-H4, pET15b-H2A, pET15b-H2B, pGEM-T-Easy-(Widom 601 145 base-pair) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Detector mode: COUNTING / Digitization - Frames/image: 1-40 / Number grids imaged: 1 / Number real images: 2408 / Average exposure time: 10.0 sec. / Average electron dose: 1.4 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: RIGID BODY FIT |
|---|
 Movie
Movie Controller
Controller
























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)