[English] 日本語
 Yorodumi
Yorodumi- EMDB-31753: Cryo-EM structure of Patched in lipid nanodisc - the wildtype, 3.... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-31753 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of Patched in lipid nanodisc - the wildtype, 3.5 angstrom (re-processed with dataset of 7dzq) | |||||||||||||||
 Map data Map data | ||||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | Caveolae / Hedgehog signaling / Lipid nanodisc / Patched / Ptc1 dimer / membrane protein | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationLigand-receptor interactions / Activation of SMO / neural plate axis specification / response to chlorate / cell differentiation involved in kidney development / hedgehog receptor activity / cell proliferation involved in metanephros development / neural tube formation / smoothened binding / hedgehog family protein binding ...Ligand-receptor interactions / Activation of SMO / neural plate axis specification / response to chlorate / cell differentiation involved in kidney development / hedgehog receptor activity / cell proliferation involved in metanephros development / neural tube formation / smoothened binding / hedgehog family protein binding / hindlimb morphogenesis / Hedgehog 'on' state / epidermal cell fate specification / spinal cord motor neuron differentiation / prostate gland development / Hedgehog 'off' state / patched binding / negative regulation of cell division / somite development / smooth muscle tissue development / pharyngeal system development / mammary gland duct morphogenesis / mammary gland epithelial cell differentiation / cellular response to cholesterol / pattern specification process / commissural neuron axon guidance / cell fate determination / metanephric collecting duct development / mammary gland development / dorsal/ventral pattern formation / regulation of growth / embryonic limb morphogenesis / dorsal/ventral neural tube patterning / negative regulation of multicellular organism growth / branching involved in ureteric bud morphogenesis / cholesterol binding / positive regulation of epidermal cell differentiation / dendritic growth cone / keratinocyte proliferation / positive regulation of cholesterol efflux / epidermis development / spermatid development / negative regulation of keratinocyte proliferation / embryonic organ development / response to retinoic acid / negative regulation of osteoblast differentiation / response to mechanical stimulus / axonal growth cone / heart morphogenesis / negative regulation of stem cell proliferation / liver regeneration / regulation of mitotic cell cycle / cyclin binding / epithelial cell proliferation / animal organ morphogenesis / stem cell proliferation / protein localization to plasma membrane / negative regulation of smoothened signaling pathway / neural tube closure / protein processing / brain development / caveola / apical part of cell / negative regulation of epithelial cell proliferation / response to estradiol / regulation of cell population proliferation / glucose homeostasis / heparin binding / regulation of protein localization / midbody / in utero embryonic development / postsynaptic membrane / cilium / response to xenobiotic stimulus / negative regulation of cell population proliferation / negative regulation of DNA-templated transcription / positive regulation of DNA-templated transcription / protein-containing complex binding / perinuclear region of cytoplasm / negative regulation of transcription by RNA polymerase II / Golgi apparatus / signal transduction / extracellular region / zinc ion binding / plasma membrane Similarity search - Function | |||||||||||||||
| Biological species |  | |||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.5 Å | |||||||||||||||
 Authors Authors | Luo Y / Zhao Y | |||||||||||||||
| Funding support |  China, 4 items China, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Structure / Year: 2021 Journal: Structure / Year: 2021Title: Cryo-EM study of patched in lipid nanodisc suggests a structural basis for its clustering in caveolae. Authors: Yitian Luo / Guoyue Wan / Xiang Zhang / Xuan Zhou / Qiuwen Wang / Jialin Fan / Hongmin Cai / Liya Ma / Hailong Wu / Qianhui Qu / Yao Cong / Yun Zhao / Dianfan Li /  Abstract: The 12-transmembrane protein Patched (Ptc1) acts as a suppressor for Hedgehog (Hh) signaling by depleting sterols in the cytoplasmic membrane leaflet that are required for the activation of ...The 12-transmembrane protein Patched (Ptc1) acts as a suppressor for Hedgehog (Hh) signaling by depleting sterols in the cytoplasmic membrane leaflet that are required for the activation of downstream regulators. The positive modulator Hh inhibits Ptc1's transporter function by binding to Ptc1 and its co-receptors, which are locally concentrated in invaginated microdomains known as caveolae. Here, we reconstitute the mouse Ptc1 into lipid nanodiscs and determine its structure using single-particle cryoelectron microscopy. The structure is overall similar to those in amphipol and detergents but displays various conformational differences in the transmembrane region. Although most particles show monomers, we observe Ptc1 dimers with distinct interaction patterns and different membrane curvatures, some of which are reminiscent of caveolae. We find that an extramembranous "hand-shake" region rich in hydrophobic and aromatic residues mediates inter-Ptc1 interactions under different membrane curvatures. Our data provide a plausible framework for Ptc1 clustering in the highly curved caveolae. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_31753.map.gz emd_31753.map.gz | 52.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-31753-v30.xml emd-31753-v30.xml emd-31753.xml emd-31753.xml | 20.5 KB 20.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_31753.png emd_31753.png | 37.9 KB | ||
| Filedesc metadata |  emd-31753.cif.gz emd-31753.cif.gz | 7.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-31753 http://ftp.pdbj.org/pub/emdb/structures/EMD-31753 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31753 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31753 | HTTPS FTP |
-Related structure data
| Related structure data |  7v6yMC  7v6zC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_31753.map.gz / Format: CCP4 / Size: 70.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_31753.map.gz / Format: CCP4 / Size: 70.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.82 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Ptc1 in lipid nanodisc
| Entire | Name: Ptc1 in lipid nanodisc |
|---|---|
| Components |
|
-Supramolecule #1: Ptc1 in lipid nanodisc
| Supramolecule | Name: Ptc1 in lipid nanodisc / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 160 kDa/nm |
-Macromolecule #1: Protein patched homolog 1,Protein patched homolog 1
| Macromolecule | Name: Protein patched homolog 1,Protein patched homolog 1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 121.637617 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MGSASAGNAA GALGRQAGGG RRRRTGGPHR AAPDRDYLHR PSYCDAAFAL EQISKGKATG RKAPLWLRAK FQRLLFKLGC YIQKNCGKF LVVGLLIFGA FAVGLKAANL ETNVEELWVE VGGRVSRELN YTRQKIGEEA MFNPQLMIQT PKEEGANVLT T EALLQHLD ...String: MGSASAGNAA GALGRQAGGG RRRRTGGPHR AAPDRDYLHR PSYCDAAFAL EQISKGKATG RKAPLWLRAK FQRLLFKLGC YIQKNCGKF LVVGLLIFGA FAVGLKAANL ETNVEELWVE VGGRVSRELN YTRQKIGEEA MFNPQLMIQT PKEEGANVLT T EALLQHLD SALQASRVHV YMYNRQWKLE HLCYKSGELI TETGYMDQII EYLYPCLIIT PLDCFWEGAK LQSGTAYLLG KP PLRWTNF DPLEFLEELK KINYQVDSWE EMLNKAEVGH GYMDRPCLNP ADPDCPATAP NKNSTKPLDV ALVLNGGCQG LSR KYMHWQ EELIVGGTVK NATGKLVSAH ALQTMFQLMT PKQMYEHFRG YDYVSHINWN EDRAAAILEA WQRTYVEVVH QSVA PNSTQ KVLPFTTTTL DDILKSFSDV SVIRVASGYL LMLAYACLTM LRWDCSKSQG AVGLAGVLLV ALSVAAGLGL CSLIG ISFN AATTQVLPFL ALGVGVDDVF LLAHAFSETG QNKRIPFEDR TGECLKRTGA SVALTSISNV TAFFMAALIP IPALRA FSL QAAVVVVFNF AMVLLIFPAI LSMDLYRRED RRLDIFCCFT SPCVSRVIQV EPQEPPCTKW TLSSFAEKHY APFLLKP KA KVVVILLFLG LLGVSLYGTT RVRDGLDLTD IVPRETREYD FIAAQFKYFS FYNMYIVTQK ADYPNIQHLL YDLHKSFS N VKYVMLEENK QLPQMWLHYF RDWLQGLQDA FDSDWETGRI MPNNYKNGSD DGVLAYKLLV QTGSRDKPID ISQLTKQRL VDADGIINPS AFYIYLTAWV SNDPVAYAAS QANIRPHRPE WVHDKADYMP ETRLRIPAAE PIEYAQFPFY LNGLRDTSDF VEAIEKVRV ICNNYTSLGL SSYPNGYPFL FWEQYISLRH WLLLSISVVL ACTFLVCAVF LLNPWTAGII VMVLALMTVE L FGMMGLIG IKLSAVPVVI LIASVGIGVE FTVHVALAFL TAIGDKNHRA MLALEHMFAP VLDGAVSTLL GVLMLAGSEF DF IVRYFFA VLAILTVLGV LNGLVLLPVL LSFFGPCPEV SPANGTLEVL FQG UniProtKB: Protein patched homolog 1, Protein patched homolog 1 |
-Macromolecule #3: CHOLESTEROL
| Macromolecule | Name: CHOLESTEROL / type: ligand / ID: 3 / Number of copies: 2 / Formula: CLR |
|---|---|
| Molecular weight | Theoretical: 386.654 Da |
| Chemical component information |  ChemComp-CLR: |
-Macromolecule #4: (2S)-2-azanyl-3-[[(2S)-3-butanoyloxy-2-dec-9-enoyloxy-propoxy]-ox...
| Macromolecule | Name: (2S)-2-azanyl-3-[[(2S)-3-butanoyloxy-2-dec-9-enoyloxy-propoxy]-oxidanyl-phosphoryl]oxy-propanoic acid type: ligand / ID: 4 / Number of copies: 1 / Formula: 5VI |
|---|---|
| Molecular weight | Theoretical: 481.474 Da |
| Chemical component information | 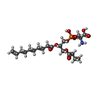 ChemComp-5VI: |
-Macromolecule #5: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 5 / Number of copies: 3 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 8.5 mg/mL | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Digitization - Dimensions - Width: 5760 pixel / Digitization - Dimensions - Height: 4092 pixel / Number grids imaged: 1 / Average exposure time: 2.23 sec. / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Calibrated defocus max: 3.0 µm / Calibrated defocus min: 1.1 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.2 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 120000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Source name: PDB / Chain - Initial model type: experimental model |
|---|---|
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT / Overall B value: 57.04 |
| Output model |  PDB-7v6y: |
 Movie
Movie Controller
Controller


















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)






















