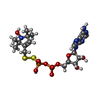
| Entry | Database: PDB / ID: 6mi6
|
|---|
| Title | STRUCTURE OF CHEA DOMAIN P4 IN COMPLEX WITH AN ADP ANALOG |
|---|
 Components Components | Chemotaxis protein CheA |
|---|
 Keywords Keywords | SIGNALING PROTEIN / bacterial chemotaxis / histidine kinase / Thermotoga maritima |
|---|
| Function / homology |  Function and homology information Function and homology information
phosphorelay sensor kinase activity / histidine kinase / chemotaxis / protein domain specific binding / ATP binding / cytoplasmSimilarity search - Function Chemotaxis protein CheA, P2 response regulator-binding / Chemotaxis protein CheA, P2 response regulator-binding domain superfamily / P2 response regulator binding domain / Histidine kinase CheA-like, homodimeric domain / CheY-binding domain of CheA / Histidine kinase CheA-like, homodimeric domain superfamily / Signal transducing histidine kinase, homodimeric domain / Signal transducing histidine kinase, homodimeric domain / : / CheW-like domain profile. ...Chemotaxis protein CheA, P2 response regulator-binding / Chemotaxis protein CheA, P2 response regulator-binding domain superfamily / P2 response regulator binding domain / Histidine kinase CheA-like, homodimeric domain / CheY-binding domain of CheA / Histidine kinase CheA-like, homodimeric domain superfamily / Signal transducing histidine kinase, homodimeric domain / Signal transducing histidine kinase, homodimeric domain / : / CheW-like domain profile. / CheW-like domain / CheW-like domain superfamily / CheW-like domain / Two component signalling adaptor domain / Histidine Phosphotransfer domain / Hpt domain / Histidine-containing phosphotransfer (HPt) domain profile. / Signal transduction histidine kinase, phosphotransfer (Hpt) domain / HPT domain superfamily / Signal transduction histidine kinase-related protein, C-terminal / Signal transduction histidine kinase, dimerisation/phosphoacceptor domain superfamily / Histidine kinase domain / Histidine kinase domain profile. / Histidine kinase-, DNA gyrase B-, and HSP90-like ATPase / Histidine kinase-like ATPases / Histidine kinase/HSP90-like ATPase superfamilySimilarity search - Domain/homology |
|---|
| Biological species |   Thermotoga maritima (bacteria) Thermotoga maritima (bacteria) |
|---|
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / Resolution: 2.95 Å SYNCHROTRON / Resolution: 2.95 Å |
|---|
 Authors Authors | Crane, B.R. / Muok, A.R. / Chua, T.K. / Le, H. |
|---|
 Citation Citation |  Journal: Appl.Magn.Reson. / Year: 2018 Journal: Appl.Magn.Reson. / Year: 2018
Title: Nucleotide Spin Labeling for ESR Spectroscopy of ATP-Binding Proteins.
Authors: Muok, A.R. / Chua, T.K. / Le, H. / Crane, B.R. |
|---|
| History | | Deposition | Sep 19, 2018 | Deposition site: RCSB / Processing site: RCSB |
|---|
| Revision 1.0 | Dec 5, 2018 | Provider: repository / Type: Initial release |
|---|
| Revision 1.1 | Nov 13, 2019 | Group: Database references / Category: citation / citation_author
Item: _citation.pdbx_database_id_PubMed / _citation.title / _citation_author.identifier_ORCID |
|---|
| Revision 1.2 | Dec 25, 2024 | Group: Advisory / Data collection ...Advisory / Data collection / Database references / Derived calculations / Structure summary
Category: chem_comp_atom / chem_comp_bond ...chem_comp_atom / chem_comp_bond / database_2 / pdbx_entry_details / pdbx_validate_close_contact / struct_conn
Item: _database_2.pdbx_DOI / _database_2.pdbx_database_accession |
|---|
|
|---|
 Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information
 Thermotoga maritima (bacteria)
Thermotoga maritima (bacteria) X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON / Resolution: 2.95 Å
SYNCHROTRON / Resolution: 2.95 Å  Authors
Authors Citation
Citation Journal: Appl.Magn.Reson. / Year: 2018
Journal: Appl.Magn.Reson. / Year: 2018 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 6mi6.cif.gz
6mi6.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb6mi6.ent.gz
pdb6mi6.ent.gz PDB format
PDB format 6mi6.json.gz
6mi6.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/mi/6mi6
https://data.pdbj.org/pub/pdb/validation_reports/mi/6mi6 ftp://data.pdbj.org/pub/pdb/validation_reports/mi/6mi6
ftp://data.pdbj.org/pub/pdb/validation_reports/mi/6mi6 Links
Links Assembly
Assembly




 Components
Components
 Thermotoga maritima (strain ATCC 43589 / MSB8 / DSM 3109 / JCM 10099) (bacteria)
Thermotoga maritima (strain ATCC 43589 / MSB8 / DSM 3109 / JCM 10099) (bacteria)
 X-RAY DIFFRACTION / Number of used crystals: 1
X-RAY DIFFRACTION / Number of used crystals: 1  Sample preparation
Sample preparation SYNCHROTRON / Site:
SYNCHROTRON / Site:  APS
APS  / Beamline: 24-ID-E / Wavelength: 0.9722 Å
/ Beamline: 24-ID-E / Wavelength: 0.9722 Å Processing
Processing Movie
Movie Controller
Controller












 PDBj
PDBj