+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-23036 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of Yeast TFIIK (Kin28/Ccl1/Tfb3) Complex | |||||||||
 Map data Map data | CryoEM structure of Yeast TFIIK (Kin28/Ccl1/Tfb3) Complex | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Polymerase CTD / TFIIH / Phosphorylation / Kinase / CDK / Cyclin / TRANSCRIPTION-Transferase complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationpositive regulation of Atg1/ULK1 kinase complex assembly / transcription factor TFIIK complex / transcription factor TFIIH holo complex / cyclin-dependent protein serine/threonine kinase activator activity / [RNA-polymerase]-subunit kinase / cellular response to nitrogen starvation / cyclin-dependent protein serine/threonine kinase regulator activity / RNA Pol II CTD phosphorylation and interaction with CE / Formation of the Early Elongation Complex / mRNA Capping ...positive regulation of Atg1/ULK1 kinase complex assembly / transcription factor TFIIK complex / transcription factor TFIIH holo complex / cyclin-dependent protein serine/threonine kinase activator activity / [RNA-polymerase]-subunit kinase / cellular response to nitrogen starvation / cyclin-dependent protein serine/threonine kinase regulator activity / RNA Pol II CTD phosphorylation and interaction with CE / Formation of the Early Elongation Complex / mRNA Capping / RNA polymerase II transcribes snRNA genes / TP53 Regulates Transcription of DNA Repair Genes / RNA Polymerase II Promoter Escape / RNA Polymerase II Transcription Pre-Initiation And Promoter Opening / RNA Polymerase II Transcription Initiation / RNA Polymerase II Transcription Initiation And Promoter Clearance / RNA Polymerase II Pre-transcription Events / Formation of TC-NER Pre-Incision Complex / RNA Polymerase I Promoter Escape / Gap-filling DNA repair synthesis and ligation in TC-NER / Dual incision in TC-NER / 7-methylguanosine mRNA capping / cyclin-dependent protein serine/threonine kinase activity / transcription by RNA polymerase I / positive regulation of autophagy / RNA polymerase II CTD heptapeptide repeat kinase activity / nucleotide-excision repair / transcription initiation at RNA polymerase II promoter / positive regulation of transcription elongation by RNA polymerase II / transcription by RNA polymerase II / protein kinase activity / regulation of cell cycle / cell division / DNA repair / regulation of transcription by RNA polymerase II / positive regulation of transcription by RNA polymerase II / mitochondrion / zinc ion binding / ATP binding / nucleus / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |   | |||||||||
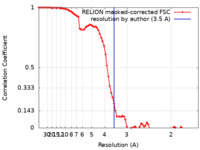
| Method | single particle reconstruction / cryo EM / Resolution: 3.5 Å | |||||||||
 Authors Authors | van Eeuwen T / Murakami K | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2021 Journal: Sci Adv / Year: 2021Title: Structure of TFIIK for phosphorylation of CTD of RNA polymerase II. Authors: Trevor van Eeuwen / Tao Li / Hee Jong Kim / Jose J Gorbea Colón / Mitchell I Parker / Roland L Dunbrack / Benjamin A Garcia / Kuang-Lei Tsai / Kenji Murakami /  Abstract: During transcription initiation, the general transcription factor TFIIH marks RNA polymerase II by phosphorylating Ser5 of the carboxyl-terminal domain (CTD) of Rpb1, which is followed by extensive ...During transcription initiation, the general transcription factor TFIIH marks RNA polymerase II by phosphorylating Ser5 of the carboxyl-terminal domain (CTD) of Rpb1, which is followed by extensive modifications coupled to transcription elongation, mRNA processing, and histone dynamics. We have determined a 3.5-Å resolution cryo-electron microscopy (cryo-EM) structure of the TFIIH kinase module (TFIIK in yeast), which is composed of Kin28, Ccl1, and Tfb3, yeast homologs of CDK7, cyclin H, and MAT1, respectively. The carboxyl-terminal region of Tfb3 was lying at the edge of catalytic cleft of Kin28, where a conserved Tfb3 helix served to stabilize the activation loop in its active conformation. By combining the structure of TFIIK with the previous cryo-EM structure of the preinitiation complex, we extend the previously proposed model of the CTD path to the active site of TFIIK. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_23036.map.gz emd_23036.map.gz | 38.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-23036-v30.xml emd-23036-v30.xml emd-23036.xml emd-23036.xml | 24.5 KB 24.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_23036_fsc.xml emd_23036_fsc.xml | 8.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_23036.png emd_23036.png | 144.7 KB | ||
| Filedesc metadata |  emd-23036.cif.gz emd-23036.cif.gz | 7 KB | ||
| Others |  emd_23036_additional_1.map.gz emd_23036_additional_1.map.gz emd_23036_half_map_1.map.gz emd_23036_half_map_1.map.gz emd_23036_half_map_2.map.gz emd_23036_half_map_2.map.gz | 3.2 MB 33 MB 33 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-23036 http://ftp.pdbj.org/pub/emdb/structures/EMD-23036 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23036 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23036 | HTTPS FTP |
-Validation report
| Summary document |  emd_23036_validation.pdf.gz emd_23036_validation.pdf.gz | 611.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_23036_full_validation.pdf.gz emd_23036_full_validation.pdf.gz | 611 KB | Display | |
| Data in XML |  emd_23036_validation.xml.gz emd_23036_validation.xml.gz | 13.5 KB | Display | |
| Data in CIF |  emd_23036_validation.cif.gz emd_23036_validation.cif.gz | 19.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23036 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23036 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23036 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23036 | HTTPS FTP |
-Related structure data
| Related structure data |  7kueMC  6xi8C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_23036.map.gz / Format: CCP4 / Size: 42.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_23036.map.gz / Format: CCP4 / Size: 42.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | CryoEM structure of Yeast TFIIK (Kin28/Ccl1/Tfb3) Complex | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.83 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: Additional map
| File | emd_23036_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Additional map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half-map 1
| File | emd_23036_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half-map 2
| File | emd_23036_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Ternary complex of Kin28-Ccl1-Tfb3 from Saccharomyces cerevisiae.
| Entire | Name: Ternary complex of Kin28-Ccl1-Tfb3 from Saccharomyces cerevisiae. |
|---|---|
| Components |
|
-Supramolecule #1: Ternary complex of Kin28-Ccl1-Tfb3 from Saccharomyces cerevisiae.
| Supramolecule | Name: Ternary complex of Kin28-Ccl1-Tfb3 from Saccharomyces cerevisiae. type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 Details: the yeast Cdk7 complex, that phosphorylates the RNA pol II C-terminal domain (CTD) in transcription initiation. |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 73 KDa |
-Macromolecule #1: RNA polymerase II transcription factor B subunit 3
| Macromolecule | Name: RNA polymerase II transcription factor B subunit 3 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 38.128355 KDa |
| Sequence | String: MLMDEYEENK DMCPICKTDR YLSPDVKFLV NPECYHRICE SCVDRIFSLG PAQCPYKGCD KILRKNKFKT QIFDDVEVEK EVDIRKRVF NVFNKTIDDF NGDLVEYNKY LEEVEDIIYK LDHGIDVAKT EEKLRTYEEL NKQLIMNNLE RSRTEIESFE Q RQKFEKEM ...String: MLMDEYEENK DMCPICKTDR YLSPDVKFLV NPECYHRICE SCVDRIFSLG PAQCPYKGCD KILRKNKFKT QIFDDVEVEK EVDIRKRVF NVFNKTIDDF NGDLVEYNKY LEEVEDIIYK LDHGIDVAKT EEKLRTYEEL NKQLIMNNLE RSRTEIESFE Q RQKFEKEM KLKKRLLERQ IEEEERMNKE WTKKEIVNRL STTTQDINET IEGVKNTVKL KKSSARRKLE ELNRVLKNNP YF NSNVNVQ NSRLKDAVPF TPFNGDREAH PPFTLKGSVY NDPFIKDLEH RKEFIASGFN TNYAYERVLT EAFMGLGCVI SEE L UniProtKB: RNA polymerase II transcription factor B subunit 3 |
-Macromolecule #2: Serine/threonine-protein kinase KIN28
| Macromolecule | Name: Serine/threonine-protein kinase KIN28 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO / EC number: [RNA-polymerase]-subunit kinase |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 35.369867 KDa |
| Sequence | String: MKVNMEYTKE KKVGEGTYAV VYLGCQHSTG RKIAIKEIKT SEFKDGLDMS AIREVKYLQE MQHPNVIELI DIFMAYDNLN LVLEFLPTD LEVVIKDKSI LFTPADIKAW MLMTLRGVYH CHRNFILHRD LKPNNLLFSP DGQIKVADFG LARAIPAPHE I L(TPO)SNVVTR ...String: MKVNMEYTKE KKVGEGTYAV VYLGCQHSTG RKIAIKEIKT SEFKDGLDMS AIREVKYLQE MQHPNVIELI DIFMAYDNLN LVLEFLPTD LEVVIKDKSI LFTPADIKAW MLMTLRGVYH CHRNFILHRD LKPNNLLFSP DGQIKVADFG LARAIPAPHE I L(TPO)SNVVTR WYRAPELLFG AKHYTSAIDI WSVGVIFAEL MLRIPYLPGQ NDVDQMEVTF RALGTPTDRD WPEVSSFM T YNKLQIYPPP SRDELRKRFI AASEYALDFM CGMLTMNPQK RWTAVQCLES DYFKELPPPS DPSSIKIRN UniProtKB: Serine/threonine-protein kinase KIN28 |
-Macromolecule #3: Cyclin CCL1
| Macromolecule | Name: Cyclin CCL1 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 45.259414 KDa |
| Sequence | String: MTDIQLNGKS TLDTPSATMS AKEKEAKLKS ADENNKPPNY KRISDSQLYR HSSQYRMWSY TKDQLQEKRV DTNARAIAYI EENLLKFRE AHNLTEEEIK VLEAKAIPLT MEEELDLVNF YAKKVQVIAQ HLNLPTEVVA TAISFFRRFF LENSVMQIDP K SIVHTTIF ...String: MTDIQLNGKS TLDTPSATMS AKEKEAKLKS ADENNKPPNY KRISDSQLYR HSSQYRMWSY TKDQLQEKRV DTNARAIAYI EENLLKFRE AHNLTEEEIK VLEAKAIPLT MEEELDLVNF YAKKVQVIAQ HLNLPTEVVA TAISFFRRFF LENSVMQIDP K SIVHTTIF LACKSENYFI SVDSFAQKAK STRDSVLKFE FKLLESLKFS LLNHHPYKPL HGFFLDIQNV LYGKVDLNYM GQ IYDRCKK RITAALLTDV VYFYTPPQIT LATLLIEDEA LVTRYLETKF PSREGSQESV PGNEKEEPQN DASTTEKNKE KST ESEEYS IDSAKLLTII RECKSIIEDC KPPSTEEAKK IAAKNYYCQN PSTLIQKLKR KLNGEDTSST VEKKQKT UniProtKB: Cyclin CCL1 |
-Macromolecule #4: ADENOSINE-5'-DIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-DIPHOSPHATE / type: ligand / ID: 4 / Number of copies: 1 / Formula: ADP |
|---|---|
| Molecular weight | Theoretical: 427.201 Da |
| Chemical component information |  ChemComp-ADP: |
-Macromolecule #5: ALUMINUM FLUORIDE
| Macromolecule | Name: ALUMINUM FLUORIDE / type: ligand / ID: 5 / Number of copies: 1 / Formula: AF3 |
|---|---|
| Molecular weight | Theoretical: 83.977 Da |
| Chemical component information |  ChemComp-AF3: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.08 mg/mL | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.6 Component:
| ||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 0 % / Chamber temperature: 293 K / Instrument: LEICA EM CPC Details: blotted for 2 seconds with Whatman 41 ashless filter paper. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number grids imaged: 1 / Number real images: 4620 / Average exposure time: 2.24 sec. / Average electron dose: 45.0 e/Å2 Details: Images collected in super-resolution mode. Movies were 35 frames. Imaging 1 image/hole, image shift between 4 holes. Focus once per image shift |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.8000000000000003 µm / Nominal defocus min: 0.6 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller
























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)