+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-21646 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | IL-23 receptor complex | |||||||||
 Map data Map data | IL-23 receptor complex | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
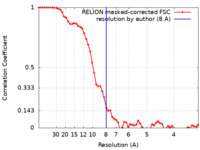
| Method | single particle reconstruction / cryo EM / Resolution: 8.0 Å | |||||||||
 Authors Authors | Glassman CR / Mathiharan YK / Panova O / Skiniotis G / Garcia KC | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Cell / Year: 2021 Journal: Cell / Year: 2021Title: Structural basis for IL-12 and IL-23 receptor sharing reveals a gateway for shaping actions on T versus NK cells. Authors: Caleb R Glassman / Yamuna Kalyani Mathiharan / Kevin M Jude / Leon Su / Ouliana Panova / Patrick J Lupardus / Jamie B Spangler / Lauren K Ely / Christoph Thomas / Georgios Skiniotis / K Christopher Garcia /  Abstract: Interleukin-12 (IL-12) and IL-23 are heterodimeric cytokines that are produced by antigen-presenting cells to regulate the activation and differentiation of lymphocytes, and they share IL-12Rβ1 as a ...Interleukin-12 (IL-12) and IL-23 are heterodimeric cytokines that are produced by antigen-presenting cells to regulate the activation and differentiation of lymphocytes, and they share IL-12Rβ1 as a receptor signaling subunit. We present a crystal structure of the quaternary IL-23 (IL-23p19/p40)/IL-23R/IL-12Rβ1 complex, together with cryoelectron microscopy (cryo-EM) maps of the complete IL-12 (IL-12p35/p40)/IL-12Rβ2/IL-12Rβ1 and IL-23 receptor (IL-23R) complexes, which reveal "non-canonical" topologies where IL-12Rβ1 directly engages the common p40 subunit. We targeted the shared IL-12Rβ1/p40 interface to design a panel of IL-12 partial agonists that preserved interferon gamma (IFNγ) induction by CD8 T cells but impaired cytokine production from natural killer (NK) cells in vitro. These cell-biased properties were recapitulated in vivo, where IL-12 partial agonists elicited anti-tumor immunity to MC-38 murine adenocarcinoma absent the NK-cell-mediated toxicity seen with wild-type IL-12. Thus, the structural mechanism of receptor sharing used by IL-12 family cytokines provides a protein interface blueprint for tuning this cytokine axis for therapeutics. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_21646.map.gz emd_21646.map.gz | 14.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-21646-v30.xml emd-21646-v30.xml emd-21646.xml emd-21646.xml | 15 KB 15 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_21646_fsc.xml emd_21646_fsc.xml | 5.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_21646.png emd_21646.png | 32.7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-21646 http://ftp.pdbj.org/pub/emdb/structures/EMD-21646 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21646 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21646 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_21646.map.gz / Format: CCP4 / Size: 15.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_21646.map.gz / Format: CCP4 / Size: 15.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | IL-23 receptor complex | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.7 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : IL-23 receptor extracellular domain complex
| Entire | Name: IL-23 receptor extracellular domain complex |
|---|---|
| Components |
|
-Supramolecule #1: IL-23 receptor extracellular domain complex
| Supramolecule | Name: IL-23 receptor extracellular domain complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: IL-23 (IL-23p19,p40), IL-12Rb1 (with affinity enhancing Y109S and Q132L mutations), IL-23R |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Molecular weight | Theoretical: 153 kDa/nm |
-Macromolecule #1: Interleukin-23 receptor
| Macromolecule | Name: Interleukin-23 receptor / type: protein_or_peptide / ID: 1 Details: hIL-23R (25-309), 3C protease site, Protein C tag, 8xHis Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: ITNINCSGHI WVEPATIFKM GMNISIYCQA AIKNCQPRKL HFYKNGIKER FQITRINKTT ARLWYKNFLE PHASMYCTAE CPKHFQETLI CGKDISSGYP PDIPDEVTCV IYEYSGNMTC TWNAGKLTYI DTKYVVHVKS LETEEEQQYL TSSYINISTD SLQGGKKYLV ...String: ITNINCSGHI WVEPATIFKM GMNISIYCQA AIKNCQPRKL HFYKNGIKER FQITRINKTT ARLWYKNFLE PHASMYCTAE CPKHFQETLI CGKDISSGYP PDIPDEVTCV IYEYSGNMTC TWNAGKLTYI DTKYVVHVKS LETEEEQQYL TSSYINISTD SLQGGKKYLV WVQAANALGM EESKQLQIHL DDIVIPSAAV ISRAETINAT VPKTIIYWDS QTTIEKVSCE MRYKATTNQT WNVKEFDTNF TYVQQSEFYL EPNIKYVFQV RCQETGKRYW QPWSSPFFHK TAAALEVLFQ GPGAAEDQVD PRLIDGKHHH HHHHH |
-Macromolecule #2: Interleukin-23 subunit alpha
| Macromolecule | Name: Interleukin-23 subunit alpha / type: protein_or_peptide / ID: 2 / Details: IL23A (28-189) / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: PAWTQCQQLS QKLCTLAWSA HPLVGHMDLR EEGDEETTND VPHIQCGDGC DPQGLRDNSQ FCLQRIHQGL IFYEKLLGSD IFTGEPSLLP DSPVGQLHAS LLGLSQLLQP EGHHWETQQI PSLSPSQPWQ RLLLRFKILR SLQAFVAVAA RVFAHGAATL SP |
-Macromolecule #3: Interleukin-12 subunit beta
| Macromolecule | Name: Interleukin-12 subunit beta / type: protein_or_peptide / ID: 3 / Details: IL12B (23-328), Biotin Acceptor Peptide, 6xHis / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: IWELKKDVYV VELDWYPDAP GEMVVLTCDT PEEDGITWTL DQSSEVLGSG KTLTIQVKEF GDAGQYTCHK GGEVLSHSLL LLHKKEDGIW STDILKDQKE PKNKTFLRCE AKNYSGRFTC WWLTTISTDL TFSVKSSRGS SDPQGVTCGA ATLSAERVRG DNKEYEYSVE ...String: IWELKKDVYV VELDWYPDAP GEMVVLTCDT PEEDGITWTL DQSSEVLGSG KTLTIQVKEF GDAGQYTCHK GGEVLSHSLL LLHKKEDGIW STDILKDQKE PKNKTFLRCE AKNYSGRFTC WWLTTISTDL TFSVKSSRGS SDPQGVTCGA ATLSAERVRG DNKEYEYSVE CQEDSACPAA EESLPIEVMV DAVHKLKYEN YTSSFFIRDI IKPDPPKNLQ LKPLKNSRQV EVSWEYPDTW STPHSYFSLT FCVQVQGKSK REKKDRVFTD KTSATVICRK NASISVRAQD RYYSSSWSEW ASVPCSGRLH HILDAQKMVW NHRHHHHHH |
-Macromolecule #4: Interleukin-12 receptor subunit beta-1
| Macromolecule | Name: Interleukin-12 receptor subunit beta-1 / type: protein_or_peptide / ID: 4 / Details: IL12RB1 (25-542) Y109S Q132L, 6xHis / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: RTSECCFQDP PYPDADSGSA SGPRDLRCYR ISSDRYECSW QYEGPTAGVS HFLRCCLSSG RCCYFAAGSA TRLQFSDQAG VSVLSTVTLW VESWARNQTE KSPEVTLLLY NSVKYEPPLG DIKVSKLAGQ LRMEWETPDN QVGAEVQFRH RTPSSPWKLG DCGPQDDDTE ...String: RTSECCFQDP PYPDADSGSA SGPRDLRCYR ISSDRYECSW QYEGPTAGVS HFLRCCLSSG RCCYFAAGSA TRLQFSDQAG VSVLSTVTLW VESWARNQTE KSPEVTLLLY NSVKYEPPLG DIKVSKLAGQ LRMEWETPDN QVGAEVQFRH RTPSSPWKLG DCGPQDDDTE SCLCPLEMNV AQEFQLRRRQ LGSQGSSWSK WSSPVCVPPE NPPQPQVRFS VEQLGQDGRR RLTLKEQPTQ LELPEGCQGL APGTEVTYRL QLHMLSCPCK AKATRTLHLG KMPYLSGAAY NVAVISSNQF GPGLNQTWHI PADTHTEPVA LNISVGTNGT TMYWPARAQS MTYCIEWQPV GQDGGLATCS LTAPQDPDPA GMATYSWSRE SGAMGQEKCY YITIFASAHP EKLTLWSTVL STYHFGGNAS AAGTPHHVSV KNHSLDSVSV DWAPSLLSTC PGVLKEYVVR CRDEDSKQVS EHPVQPTETQ VTLSGLRAGV AYTVQVRADT AWLRGVWSQP QRFSIEVQAA AHHHHHH |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.04 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
| ||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 200 / Pretreatment - Type: GLOW DISCHARGE | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 293 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 44.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.4 µm / Nominal defocus min: 1.5 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)