+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-21617 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of a substrate-bound DQC ubiquitin ligase | |||||||||
 Map data Map data | Unfiltered half-map from RELION auto-refinement | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | ubiquitin / E3-ligase / multiprotein complex / substrate recognition / LIGASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationParkin-FBXW7-Cul1 ubiquitin ligase complex / F-box domain binding / PcG protein complex / regulation of epidermal cell differentiation / cullin-RING ubiquitin ligase complex / positive regulation of ubiquitin protein ligase activity / Cul7-RING ubiquitin ligase complex / maintenance of protein location in nucleus / Loss of Function of FBXW7 in Cancer and NOTCH1 Signaling / regulation of smoothened signaling pathway ...Parkin-FBXW7-Cul1 ubiquitin ligase complex / F-box domain binding / PcG protein complex / regulation of epidermal cell differentiation / cullin-RING ubiquitin ligase complex / positive regulation of ubiquitin protein ligase activity / Cul7-RING ubiquitin ligase complex / maintenance of protein location in nucleus / Loss of Function of FBXW7 in Cancer and NOTCH1 Signaling / regulation of smoothened signaling pathway / Nuclear events mediated by NFE2L2 / negative regulation of response to oxidative stress / SCF ubiquitin ligase complex / neural crest cell differentiation / SCF-dependent proteasomal ubiquitin-dependent protein catabolic process / Cul3-RING ubiquitin ligase complex / Prolactin receptor signaling / ubiquitin ligase complex scaffold activity / protein quality control for misfolded or incompletely synthesized proteins / cullin family protein binding / protein monoubiquitination / ubiquitin-like ligase-substrate adaptor activity / protein K48-linked ubiquitination / transcription regulator inhibitor activity / Nuclear events stimulated by ALK signaling in cancer / inclusion body / cellular response to interleukin-4 / intrinsic apoptotic signaling pathway / molecular function activator activity / animal organ morphogenesis / Regulation of BACH1 activity / MAP3K8 (TPL2)-dependent MAPK1/3 activation / actin filament / SCF-beta-TrCP mediated degradation of Emi1 / NIK-->noncanonical NF-kB signaling / Vpu mediated degradation of CD4 / Dectin-1 mediated noncanonical NF-kB signaling / Activation of NF-kappaB in B cells / Degradation of GLI1 by the proteasome / Iron uptake and transport / G1/S transition of mitotic cell cycle / GSK3B and BTRC:CUL1-mediated-degradation of NFE2L2 / Negative regulation of NOTCH4 signaling / FBXL7 down-regulates AURKA during mitotic entry and in early mitosis / Degradation of GLI2 by the proteasome / GLI3 is processed to GLI3R by the proteasome / beta-catenin binding / Degradation of beta-catenin by the destruction complex / NOTCH1 Intracellular Domain Regulates Transcription / CLEC7A (Dectin-1) signaling / Constitutive Signaling by NOTCH1 PEST Domain Mutants / Constitutive Signaling by NOTCH1 HD+PEST Domain Mutants / SCF(Skp2)-mediated degradation of p27/p21 / FCERI mediated NF-kB activation / centriolar satellite / Interleukin-1 signaling / protein polyubiquitination / Orc1 removal from chromatin / disordered domain specific binding / Regulation of RUNX2 expression and activity / Cyclin D associated events in G1 / : / KEAP1-NFE2L2 pathway / Regulation of PLK1 Activity at G2/M Transition / Downstream TCR signaling / nervous system development / positive regulation of proteasomal ubiquitin-dependent protein catabolic process / Antigen processing: Ubiquitination & Proteasome degradation / Neddylation / cellular response to oxidative stress / ubiquitin-dependent protein catabolic process / midbody / protein-macromolecule adaptor activity / in utero embryonic development / Potential therapeutics for SARS / proteasome-mediated ubiquitin-dependent protein catabolic process / RNA polymerase II-specific DNA-binding transcription factor binding / positive regulation of canonical NF-kappaB signal transduction / cell population proliferation / Ub-specific processing proteases / regulation of autophagy / protein ubiquitination / chromatin remodeling / protein domain specific binding / ubiquitin protein ligase binding / centrosome / negative regulation of transcription by RNA polymerase II / endoplasmic reticulum / nucleoplasm / identical protein binding / nucleus / plasma membrane / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 8.5 Å | |||||||||
 Authors Authors | Mena EL / Jevtic P | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2020 Journal: Nature / Year: 2020Title: Structural basis for dimerization quality control. Authors: Elijah L Mena / Predrag Jevtić / Basil J Greber / Christine L Gee / Brandon G Lew / David Akopian / Eva Nogales / John Kuriyan / Michael Rape /  Abstract: Most quality control pathways target misfolded proteins to prevent toxic aggregation and neurodegeneration. Dimerization quality control further improves proteostasis by eliminating complexes of ...Most quality control pathways target misfolded proteins to prevent toxic aggregation and neurodegeneration. Dimerization quality control further improves proteostasis by eliminating complexes of aberrant composition, but how it detects incorrect subunits remains unknown. Here we provide structural insight into target selection by SCF-FBXL17, a dimerization-quality-control E3 ligase that ubiquitylates and helps to degrade inactive heterodimers of BTB proteins while sparing functional homodimers. We find that SCF-FBXL17 disrupts aberrant BTB dimers that fail to stabilize an intermolecular β-sheet around a highly divergent β-strand of the BTB domain. Complex dissociation allows SCF-FBXL17 to wrap around a single BTB domain, resulting in robust ubiquitylation. SCF-FBXL17 therefore probes both shape and complementarity of BTB domains, a mechanism that is well suited to establish quality control of complex composition for recurrent interaction modules. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_21617.map.gz emd_21617.map.gz | 5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-21617-v30.xml emd-21617-v30.xml emd-21617.xml emd-21617.xml | 26 KB 26 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_21617.png emd_21617.png | 91 KB | ||
| Filedesc metadata |  emd-21617.cif.gz emd-21617.cif.gz | 7.4 KB | ||
| Others |  emd_21617_half_map_1.map.gz emd_21617_half_map_1.map.gz emd_21617_half_map_2.map.gz emd_21617_half_map_2.map.gz | 4 MB 4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-21617 http://ftp.pdbj.org/pub/emdb/structures/EMD-21617 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21617 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21617 | HTTPS FTP |
-Validation report
| Summary document |  emd_21617_validation.pdf.gz emd_21617_validation.pdf.gz | 617.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_21617_full_validation.pdf.gz emd_21617_full_validation.pdf.gz | 617.4 KB | Display | |
| Data in XML |  emd_21617_validation.xml.gz emd_21617_validation.xml.gz | 7.9 KB | Display | |
| Data in CIF |  emd_21617_validation.cif.gz emd_21617_validation.cif.gz | 9.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-21617 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-21617 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-21617 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-21617 | HTTPS FTP |
-Related structure data
| Related structure data |  6wcqMC  6w66C  6w67C  6w68C  6w69C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_21617.map.gz / Format: CCP4 / Size: 5.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_21617.map.gz / Format: CCP4 / Size: 5.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unfiltered half-map from RELION auto-refinement | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.28 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
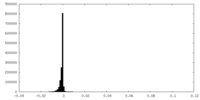
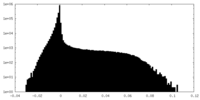
-Half map: Unfiltered half-map from RELION auto-refinement
| File | emd_21617_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unfiltered half-map from RELION auto-refinement | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
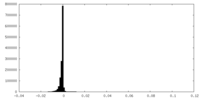
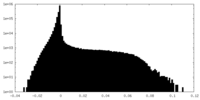
-Half map: Unfiltered half-map from RELION auto-refinement
| File | emd_21617_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unfiltered half-map from RELION auto-refinement | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : CUL1-SKP1-FBXL17-KEAP1 complex
| Entire | Name: CUL1-SKP1-FBXL17-KEAP1 complex |
|---|---|
| Components |
|
-Supramolecule #1: CUL1-SKP1-FBXL17-KEAP1 complex
| Supramolecule | Name: CUL1-SKP1-FBXL17-KEAP1 complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: S-phase kinase-associated protein 1
| Macromolecule | Name: S-phase kinase-associated protein 1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 18.679965 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MPSIKLQSSD GEIFEVDVEI AKQSVTIKTM LEDLGMDDEG DDDPVPLPNV NAAILKKVIQ WCTHHKDDPP PPEDDENKEK RTDDIPVWD QEFLKVDQGT LFELILAANY LDIKGLLDVT CKTVANMIKG KTPEEIRKTF NIKNDFTEEE EAQVRKENQW C EEK UniProtKB: S-phase kinase-associated protein 1 |
-Macromolecule #2: F-box/LRR-repeat protein 17
| Macromolecule | Name: F-box/LRR-repeat protein 17 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 44.970043 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: GEFMCHREPP PETPDINQLP PSILLKIFSN LSLDERCLSA SLVCKYWRDL CLDFQFWKQL DLSSRQQVTD ELLEKIASRS QNIIEINIS DCRSMSDNGV CVLAFKCPGL LRYTAYRCKQ LSDTSIIAVA SHCPLLQKVH VGNQDKLTDE GLKQLGSKCR E LKDIHFGQ ...String: GEFMCHREPP PETPDINQLP PSILLKIFSN LSLDERCLSA SLVCKYWRDL CLDFQFWKQL DLSSRQQVTD ELLEKIASRS QNIIEINIS DCRSMSDNGV CVLAFKCPGL LRYTAYRCKQ LSDTSIIAVA SHCPLLQKVH VGNQDKLTDE GLKQLGSKCR E LKDIHFGQ CYKISDEGMI VIAKGCLKLQ RIYMQENKLV TDQSVKAFAE HCPELQYVGF MGCSVTSKGV IHLTKLRNLS SL DLRHITE LDNETVMEIV KRCKNLSSLN LCLNWIINDR CVEVIAKEGQ NLKELYLVSC KITDYALIAI GRYSMTIETV DVG WCKEIT DQGATLIAQS SKSLRYLGLM RCDKVNEVTV EQLVQQYPHI TFSTVLQDCK RTLERAYQMG WTPNMSAASS UniProtKB: F-box/LRR-repeat protein 17 |
-Macromolecule #3: Kelch-like ECH-associated protein 1
| Macromolecule | Name: Kelch-like ECH-associated protein 1 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 70.173484 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: GGGSGGSMQP DPRPSGAGAC CRFLPLQSQC PEGAGDAVMY ASTECKAEVT PSQHGNRTFS YTLEDHTKQA FGIMNELRLS QQLCDVTLQ VKYQDAPAAQ FMAHKVALAS SSPVFKAMFT NGLREQGMEV VSIEGIHPKV MERLIEFAYT ASISMGEKCV L HVMNGAVM ...String: GGGSGGSMQP DPRPSGAGAC CRFLPLQSQC PEGAGDAVMY ASTECKAEVT PSQHGNRTFS YTLEDHTKQA FGIMNELRLS QQLCDVTLQ VKYQDAPAAQ FMAHKVALAS SSPVFKAMFT NGLREQGMEV VSIEGIHPKV MERLIEFAYT ASISMGEKCV L HVMNGAVM YQIDSVVRAC SDFLVQQLDP SNAIGIANFA EQIGCVELHQ RAREYIYMHF GEVAKQEEFF NLSHCQLVTL IS RDDLNVR CESEVFHACI NWVKYDCEQR RFYVQALLRA VRCHSLTPNF LQMQLQKCEI LQSDSRCKDY LVKIFEELTL HKP TQVMPC RAPKVGRLIY TAGGYFRQSL SYLEAYNPSD GTWLRLADLQ VPRSGLAGCV VGGLLYAVGG RNNSPDGNTD SSAL DCYNP MTNQWSPCAP MSVPRNRIGV GVIDGHIYAV GGSHGCIHHN SVERYEPERD EWHLVAPMLT RRIGVGVAVL NRLLY AVGG FDGTNRLNSA ECYYPERNEW RMITAMNTIR SGAGVCVLHN CIYAAGGYDG QDQLNSVERY DVETETWTFV APMKHR RSA LGITVHQGRI YVLGGYDGHT FLDSVECYDP DTDTWSEVTR MTSGRSGVGV AVTMEPCRKQ IDQQNCTC UniProtKB: Kelch-like ECH-associated protein 1 |
-Macromolecule #4: Cullin-1
| Macromolecule | Name: Cullin-1 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 49.92652 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MSSTRSQNPH GLKQIGLDQI WDDLRAGIQQ VYTRQSMAKS RYMELYTHVY NYCTSVHQSN QARGAGVPPS KSKKGQTPGG AQFVGLELY KRLKEFLKNY LTNLLKDGED LMDESVLKFY TQQWEDYRFS SKVLNGICAY LNRHWVRREC DEGRKGIYEI Y SLALVTWR ...String: MSSTRSQNPH GLKQIGLDQI WDDLRAGIQQ VYTRQSMAKS RYMELYTHVY NYCTSVHQSN QARGAGVPPS KSKKGQTPGG AQFVGLELY KRLKEFLKNY LTNLLKDGED LMDESVLKFY TQQWEDYRFS SKVLNGICAY LNRHWVRREC DEGRKGIYEI Y SLALVTWR DCLFRPLNKQ VTNAVLKLIE KERNGETINT RLISGVVQSY VELGLNEDDA FAKGPTLTVY KESFESQFLA DT ERFYTRE STEFLQQNPV TEYMKKAEAR LLEEQRRVQV YLHESTQDEL ARKCEQVLIE KHLEIFHTEF QNLLDADKNE DLG RMYNLV SRIQDGLGEL KKLLETHIHN QGLAAIEKCG EAALNDPKMY VQTVLDVHKK YNALVMSAFN NDAGFVAALD KACG RFINN NAVTKMAQSS SKSPELLARY CDSLLKKSS UniProtKB: Cullin-1 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation #1
Sample preparation #1
| Preparation ID | 1 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| ||||||||||||
| Grid | Model: Quantifoil, UltrAuFoil, R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Film type ID: 1 / Support film - Material: GRAPHENE OXIDE / Support film - topology: CONTINUOUS / Pretreatment - Type: GLOW DISCHARGE Details: Glow discharge before deposition of graphene oxide. | ||||||||||||
| Vitrification | Cryogen name: ETHANE-PROPANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV | ||||||||||||
| Details | Crosslinked with BS3 |
- Sample preparation #2
Sample preparation #2
| Preparation ID | 2 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| ||||||||||||
| Grid | Model: Quantifoil, UltrAuFoil, R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Film type ID: 1 / Support film - Material: GRAPHENE OXIDE / Support film - topology: CONTINUOUS / Pretreatment - Type: GLOW DISCHARGE Details: Glow discharge before deposition of graphene oxide. | ||||||||||||
| Vitrification | Cryogen name: ETHANE-PROPANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV | ||||||||||||
| Details | Crosslinked with BS3 |
- Electron microscopy #1
Electron microscopy #1
| Microscopy ID | 1 |
|---|---|
| Microscope | FEI TALOS ARCTICA |
| Image recording | Image recording ID: 1 / Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 1 / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 2.0 µm |
| Sample stage | Specimen holder model: OTHER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
- Electron microscopy #1~
Electron microscopy #1~
| Microscopy ID | 1 |
|---|---|
| Microscope | FEI TALOS ARCTICA |
| Image recording | Image recording ID: 2 / Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Number grids imaged: 1 / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.5 µm |
| Sample stage | Specimen holder model: OTHER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| ||||||
|---|---|---|---|---|---|---|---|
| Details | Coordinates were fitted as rigid bodies or fragments in Chimera and subsequently geometry-optimized using PHENIX real space refinement. | ||||||
| Refinement | Space: REAL / Protocol: OTHER | ||||||
| Output model |  PDB-6wcq: |
 Movie
Movie Controller
Controller


























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)