[English] 日本語
 Yorodumi
Yorodumi- EMDB-1489: Human Adenovirus type 5 with 31 residue extension at the N-termin... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-1489 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Human Adenovirus type 5 with 31 residue extension at the N-terminus of polypeptide IIIa | |||||||||
 Map data Map data | Human adenovirus type 5 | |||||||||
 Sample Sample |
| |||||||||
| Biological species |   Human adenovirus 5 Human adenovirus 5 | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 14.0 Å | |||||||||
 Authors Authors | San Martin C / Glasgow JN / Borovjagin A / Beatty MS / Kashentseva EA / Curiel DT / Marabini R / Dmitriev IP | |||||||||
 Citation Citation |  Journal: J Mol Biol / Year: 2008 Journal: J Mol Biol / Year: 2008Title: Localization of the N-terminus of minor coat protein IIIa in the adenovirus capsid. Authors: Carmen San Martín / Joel N Glasgow / Anton Borovjagin / Matthew S Beatty / Elena A Kashentseva / David T Curiel / Roberto Marabini / Igor P Dmitriev /  Abstract: Minor coat protein IIIa is conserved in all adenoviruses (Ads) and is required for correct viral assembly, but its precise function in capsid organization is unknown. The latest Ad capsid model ...Minor coat protein IIIa is conserved in all adenoviruses (Ads) and is required for correct viral assembly, but its precise function in capsid organization is unknown. The latest Ad capsid model proposes that IIIa is located underneath the vertex region. To obtain experimental evidence on the location of IIIa and to further define its role, we engineered the IIIa gene to encode heterologous N-terminal peptide extensions. Recombinant Ad variants with IIIa encoding six-histidine (6His) tag, 6His, and FLAG peptides, or with 6His linked to FLAG with a (Gly(4)Ser)(3) linker were rescued and analyzed for virus yield, capsid incorporation of heterologous peptides, and capsid stability. Longer extensions could not be rescued. Western blot analysis confirmed that the modified IIIa proteins were expressed in infected cells and incorporated into virions. In the Ad encoding the 6His-linker-FLAG-IIIa gene, the 6His tag was present in light particles, but not in mature virions. Immunoelectron microscopy of this virus showed that the FLAG epitope is not accessible to antibodies on the viral particles. Three-dimensional electron microscopy and difference mapping located the IIIa N-terminal extension beneath the vertex complex, wedged at the interface between the penton base and peripentonal hexons, therefore supporting the latest proposed model. The position of the IIIa N-terminus and its low tolerance for modification provide new clues for understanding the role of this minor coat protein in Ad capsid assembly and disassembly. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_1489.map.gz emd_1489.map.gz | 67.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-1489-v30.xml emd-1489-v30.xml emd-1489.xml emd-1489.xml | 8.4 KB 8.4 KB | Display Display |  EMDB header EMDB header |
| Images |  1489.gif 1489.gif | 71.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-1489 http://ftp.pdbj.org/pub/emdb/structures/EMD-1489 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1489 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1489 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_1489.map.gz / Format: CCP4 / Size: 77.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_1489.map.gz / Format: CCP4 / Size: 77.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Human adenovirus type 5 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 4.2 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
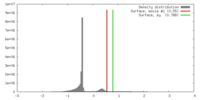
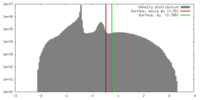
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Human adenovirus type 5
| Entire | Name:   Human adenovirus type 5 Human adenovirus type 5 |
|---|---|
| Components |
|
-Supramolecule #1000: Human adenovirus type 5
| Supramolecule | Name: Human adenovirus type 5 / type: sample / ID: 1000 / Number unique components: 1 |
|---|
-Supramolecule #1: Human adenovirus 5
| Supramolecule | Name: Human adenovirus 5 / type: virus / ID: 1 / Name.synonym: Ad5 / NCBI-ID: 28285 / Sci species name: Human adenovirus 5 / Virus type: VIRION / Virus isolate: SEROTYPE / Virus enveloped: No / Virus empty: No / Syn species name: Ad5 |
|---|---|
| Host (natural) | Organism:  Homo sapiens (human) / synonym: VERTEBRATES Homo sapiens (human) / synonym: VERTEBRATES |
| Molecular weight | Experimental: 150 MDa / Theoretical: 150 MDa |
| Virus shell | Shell ID: 1 / Diameter: 900 Å |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.2 / Details: 10 mM phosphate pH 7.2, 150 mM NaCl |
|---|---|
| Grid | Details: Quantifoil R2/4 |
| Vitrification | Cryogen name: ETHANE / Instrument: LEICA EM CPC / Details: Vitrification instrument: Leica CPC |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Image recording | Category: FILM / Film or detector model: KODAK SO-163 FILM / Digitization - Scanner: ZEISS SCAI / Digitization - Sampling interval: 7 µm / Number real images: 89 / Average electron dose: 10 e/Å2 / Bits/pixel: 8 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.2 mm / Nominal defocus max: 5.0 µm / Nominal defocus min: 1.7 µm / Nominal magnification: 50000 |
| Sample stage | Specimen holder: Eucentric / Specimen holder model: GATAN LIQUID NITROGEN |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
- Image processing
Image processing
| CTF correction | Details: phase flip in micrograph |
|---|---|
| Final reconstruction | Applied symmetry - Point group: I (icosahedral) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 14.0 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: spider,xmipp / Number images used: 3171 |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)